THE LIVING WORLD
Unit Three. The Continuity of Life
11.4. How the DNA Molecule Copies Itself
The attraction that holds the two DNA strands together is the formation of weak hydrogen bonds between the bases that face each other from the two strands. That is why A pairs with T and not C; A can only form hydrogen bonds with T. Similarly, G can form hydrogen bonds with C but not T. In the Watson-Crick model of DNA, the two strands of the double helix are said to be complementary to each other. One chain of the helix can have any sequence of bases, of A, T, G, and C, but this sequence completely determines that of its partner in the helix. If the sequence of one chain is ATTGCAT, the sequence of its partner in the double helix must be TAACGTA. Each chain in the helix is a complementary mirror image of the other. This complementarity makes it possible for the DNA molecule to copy itself during cell division in a very direct manner. But, there are three possible alternatives as to how the DNA could serve as a template for the assembly of new DNA molecules.
First, the two strands of the double helix could separate and serve as templates for the assembly of two new strands by base pairing A with T and G with C. This is what happens in figure 11.5a, with the original strand colored blue and the newly formed strands red. After replicating, the original strands rejoin, preserving the original strand of DNA and forming an entirely new strand. This is called conservative replication.
In the second alternative, the double helix need only “unzip” and assemble a new complementary chain along each single strand. This form of DNA replication is called semiconservative replication, because while the sequence of the original duplex is conserved after one round of replication, the duplex itself is not. Instead, each strand of the duplex becomes part of another duplex. You can see in figure 11.5b that the blue strand is from the original helix and the red strand is newly formed.
In the third alternative, called dispersive replication, the original DNA would serve as a template for the formation of new DNA strands but the new and old DNA would be dispersed among the two daughter strands. As shown in figure 11.5c, each daughter strand is made up of sections of original (blue) strands and new (red) strands.
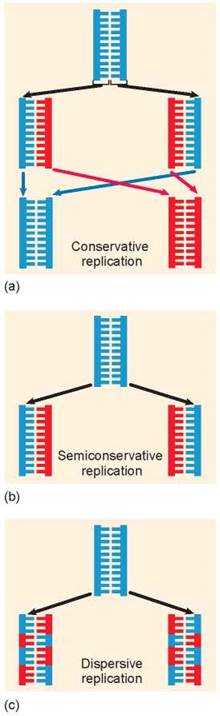
Figure 11.5. Alternative mechanisms of DNA replication.
The three alternative hypotheses of DNA replication were tested in 1958 by Matthew Meselson and Franklin Stahl of the California Institute of Technology. These two scientists grew bacteria in a medium containing the heavy isotope of nitrogen, 15N, which became incorporated into the bases of the bacterial DNA (the upper petri dish in figure 11.6). After several generations, samples were taken from this culture and grown in a medium containing the normal lighter isotope 14N, which became incorporated into the newly replicating DNA. Bacterial samples were taken from the 14N media at 20 minute intervals (2 through 4). DNA was extracted from all three samples and a fourth sample, 1, that served as a control.
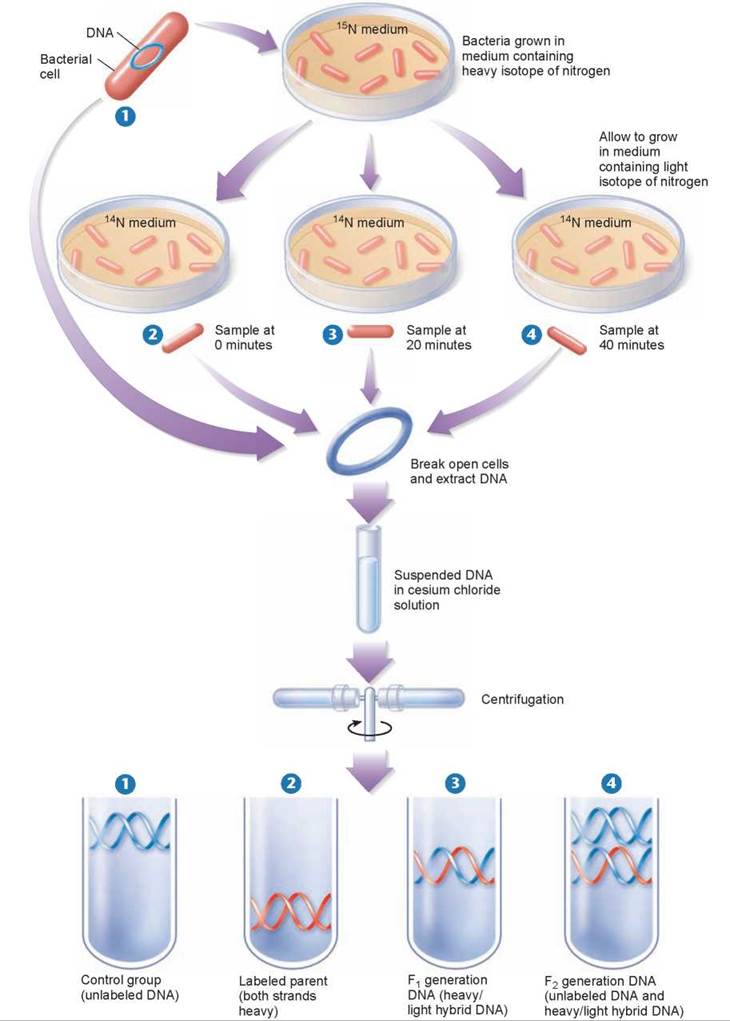
Figure 11.6. The Meselson-Stahl experiment.
Bacterial cells were grown for several generations in a medium containing a heavy isotope of nitrogen (15N) and then were transferred to a new medium containing the normal lighter isotope (14N). (The bacteria shown here are not drawn to scale, as tens of thousands of bacterial cells grow on even a tiny portion of a plate in culture.) At various times thereafter, samples of the bacteria were collected, and their DNA was dissolved in a solution of cesium chloride, which was spun rapidly in a centrifuge. The labeled and unlabeled DNA settled in different areas of the tube because they differed in weight. The DNA with two heavy strands settled down toward the bottom of the tube. The DNA with two light strands settled higher up in the tube. The DNA with one heavy and one light strand settled in between the other two.
By dissolving the DNA they had collected in a heavy salt called cesium chloride, and then spinning the solution at very high speeds in an ultracentrifuge, Meselson and Stahl were able to separate DNA strands of different densities. The centrifugal forces caused the cesium ions to migrate toward the bottom of the centrifuge tube, creating a gradient of cesium concentration, and thus a gradation of density. Each DNA strand floats or sinks in the gradient until it reaches the position where its density exactly matches the density of the cesium there. Because 15N strands are denser than 14N strands, they migrate farther down the tube to a denser region of cesium.
The DNA collected immediately after the transfer was all dense, as shown in test tube 2. However, after the bacteria completed their first round of DNA replication in the 14N medium, the density of their DNA had decreased to a value intermediate between 14N-DNA and 15N-DNA, as shown in test tube 3. After the second round of replication, two density classes of DNA were observed, one intermediate and one equal to that of 14N-DNA, as shown in test tube 4.
Meselson and Stahl interpreted their results as follows: After the first round of replication, each daughter DNA duplex was a hybrid possessing one of the heavy strands of the parent molecule and one light strand; when this hybrid duplex replicated, it contributed one heavy strand to form another hybrid duplex and one light strand to form a light duplex. Thus, this experiment clearly ruled out conservative and dispersive DNA replication, and confirmed the prediction of the Watson-Crick model that DNA replicates in a semiconservative manner.
How DNA Copies Itself
The copying of DNA before cell division is called DNA replication. This process is overseen by six proteins in prokaryotes (in eukaryotes, some of the enzymes are different). These proteins coordinate the unwinding of the DNA duplex and assembly of new complementary DNA strands by the addition of nucleotides to existing strands (figure 11.7). Here is how the process works.
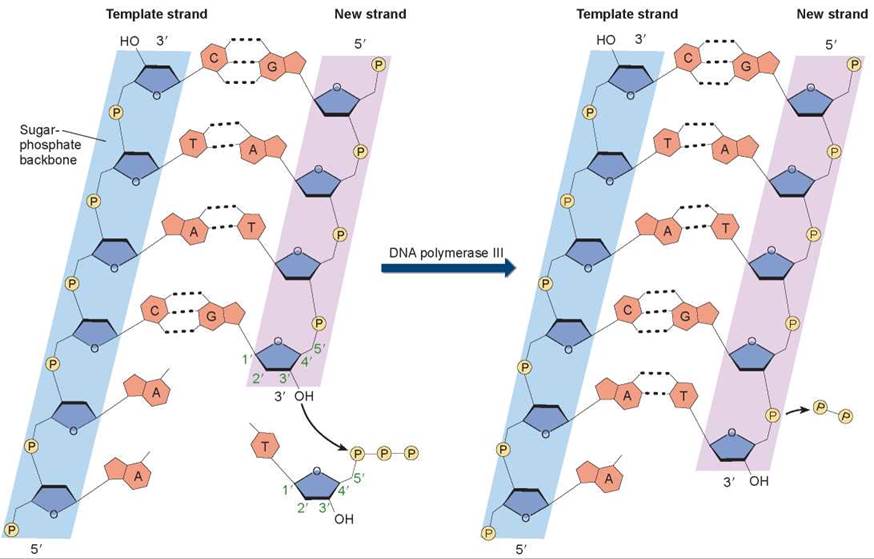
Figure 11.7. How nucleotides are added in DNA replication.
In a nucleotide, the phosphate group is attached to the 5' carbon atom of the sugar, and an OH group is attached to the 3' carbon atom. So, on a DNA strand, there will be a 5' phosphate at one end of the chain and a 3' OH at the other end. In the DNA double helix, the two strands of nucleotides pair up in opposite orientations, with one strand running 5' to 3' and the other running 3' to 5'. When nucleotides are added to a growing strand of DNA by the enzyme DNA polymerase III, the first phosphate group of the incoming nucleotide attaches to the OH group on the end nucleotide of the existing strand.
Before DNA replication can begin, an enzyme called helicase separates and unwinds the strands of the parental DNA. Single-strand binding proteins stabilize the singlestranded regions of DNA before they are replicated. Helicase moves up the DNA helix, unwinding as it goes.
After the parental DNA duplex is unwound, an enzyme complex called DNA polymerase III can add nucleotides complementary to the exposed template strand of DNA. But this enzyme cannot begin a new strand; it can only add nucleotides to an existing strand. Thus, before a new strand of DNA can be built, an enzyme called primase must synthesize a short section of joined RNA nucleotides, called a primer, complementary to the single-stranded template. DNA polymerase III can then add onto the primer and assemble a complementary new strand of DNA on each old strand. One of the new strands of DNA is called the leading strand; it is the one that is extended in a 5' to 3' direction toward the replication fork.
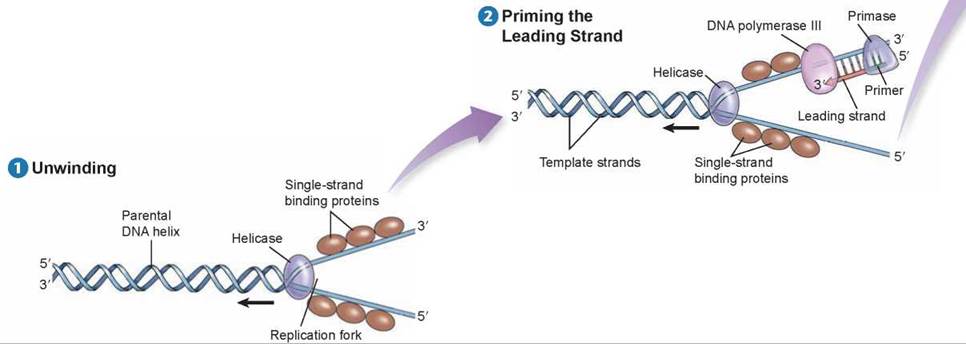
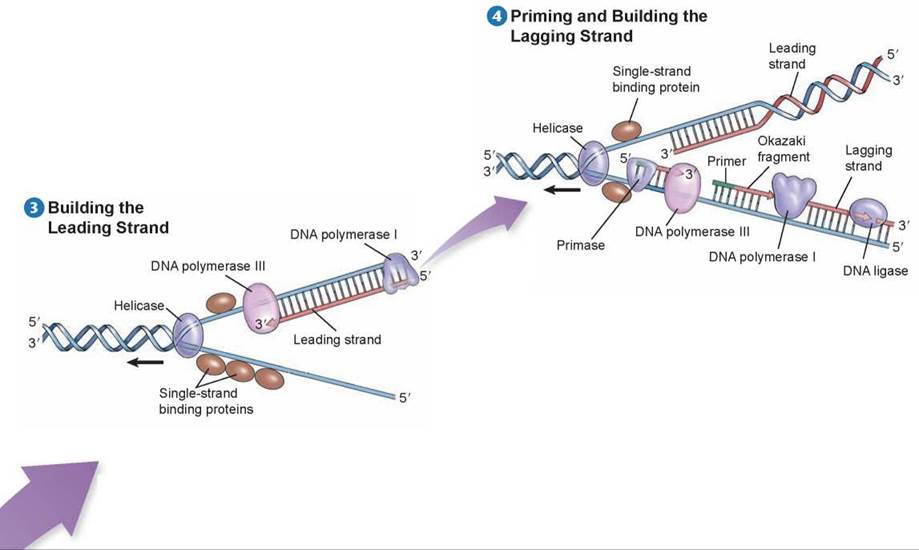
DNA polymerase III builds the leading strand by adding nucleotides to the 3’ end of the strand: the 5’ phosphate end of a nucleotide to be added attaches to the 3’ sugar end of the existing strand. DNA polymerase III moves toward the replication fork and builds the leading strand as one continuous strand. Before the leading strand can be completed, another polymerase enzyme called DNA polymerase I removes the RNA primer and fills in the gap with DNA nucleotides. The newly synthesized hybrid DNA can then rewind into a helix.
Because DNA polymerase III can only assemble new strands in the 5’ to 3’ direction, the other strand, called the lagging strand, is assembled in short 5’ to 3’ segments, moving away from the replication fork. Each lagging strand segment begins with an RNA primer, and then DNA polymerase III builds away from the replication fork until it encounters the previously synthesized section. These short stretches of newly synthesized DNA on the lagging strand are called Okazaki fragments. As the helix opens further, a new RNA primer is added, and DNA polymerase III must release the template it has completed and begin with the “new” template. The Okazaki fragments are then linked when DNA polymerase I removes the RNA primers and an enzyme called DNA ligase joins the ends of the newly synthesized segments of DNA. The entire lagging strand can only be replicated in this discontinuous fashion.
Eukaryotic chromosomes each contain a single, very long molecule of DNA (figure 11.8), one far too long to copy all the way from one end to the other with a single replication fork. Each eukaryotic chromosome is instead copied in sections of about 100,000 nucleotides, each with its own replication origin and fork.
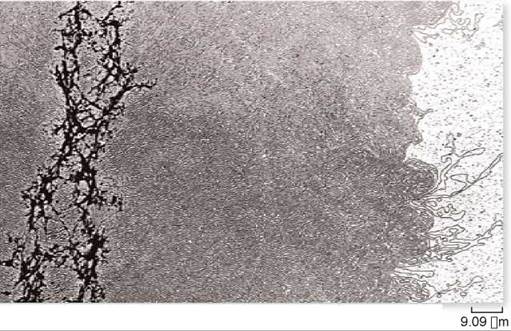
Figure 11.8. DNA of a single chromosome.
This chromosome has been relieved of most of its packaging proteins, leaving the DNA in its extended form. The residual protein scaffolding appears as the dark material on the left side of the micrograph.
The enormous amount of DNA that resides within the cells of your body represents a long series of DNA replications, starting with the DNA of a single cell—the fertilized egg. Living cells have evolved many mechanisms to avoid errors during DNA replication and to preserve the DNA from damage. These mechanisms of DNA repair proofread the strands of each daughter cell against one another for accuracy and correct any mistakes. But the proofreading is not perfect. If it were, no mistakes such as mutations would occur, no variation in gene sequence would result, and evolution would come to a halt. Mutation will be discussed in more detail in the next section and in chapter 14.
Key Learning Outcome 11.4. The basis for the great accuracy of DNA replication is complementarity. DNA's two strands are complementary mirror images of each other, so either one can be used as a template to reconstruct the other.