THE LIVING WORLD
Unit Three. The Continuity of Life

This sheep, Dolly, was the first animal to be cloned from a single adult cell. The lamb you see beside her is her offspring, normal in every respect. From Dolly we learn that genes are not lost during development. If a single adult cell can be induced to switch the proper combination of genes on and off, that one cell can develop into a normal adult individual. Embryonic stem cells are like this—poised to become any cell of the body as the embryo develops. It may be possible to replace damaged tissues with healthy tissue grown from a patient’s own embryonic stem cells, so long as the disorder is not an inherited one. The approach has been used successfully in laboratory mice to cure a variety of disorders. However, its use in humans would require employing stem cells derived from the patient, and because this may involve destroying a human embryo the approach is controversial. Another approach, when the damaged tissue does result from a defective gene, is to repair rather than replace, using a virus to transfer a healthy gene into those tissues that lack it. In this chapter you will explore genomic screening, the application of gene technology to medicine and agriculture, reproductive cloning, stem cell tissue replacement, and gene therapy, all areas in which a revolution is reshaping biology.
13.1. Genomics
Recent years have seen an explosion of interest in comparing the entire DNA content of different organisms, a new field of biology called genomics. While initial successes focused on organisms with relatively small numbers of genes, researchers have recently completed the sequencing of several large eukaryotic genomes, including our own.
The full complement of genetic information of an organism—all of its genes and other DNA—is called its genome. To study a genome, the DNA is first sequenced, a process that allows each nucleotide of a DNA strand to be read in order. The first genome to be sequenced was a very simple one: a small bacterial virus called Ø-X174 (Ø is the Greek letter phi). Frederick Sanger, inventor of the first practical way to sequence DNA, obtained the sequence of this 5,375-nucleotide genome in 1977. This was followed by the sequencing of dozens of prokaryotic genomes. The advent of automated DNA sequencing machines in recent years has made the DNA sequencing of much larger eukaryotic genomes practical, including our own (table 13.1).
TABLE 13.1. SOME EUKARYOTIC GENOMES
Organism |
Estimated Genome Size (Mbp) |
Number of Genes (x1,000) |
Nature of Genome |
|
Vertebrates |
||||
|
Homo sapiens (human) |
3,200 |
20-25 |
The first large genome to be sequenced; the number of transcribable genes is far less than expected; much of the genome is occupied by repeated DNA sequences. |
|
Pan troglodytes (chimpanzee) |
2,800 |
20-25 |
There are few base substitutions between chimp and human genomes, less than 2%, but many small sequences of DNA have been lost as the two species diverged, often with significant effects. |
|
Mas musculus (mouse) |
2,500 |
25 |
Roughly 80% of mouse genes have a functional equivalent in the human genome; importantly, large portions of the noncoding DNA of mouse and human have been conserved; overall, rodent genomes (mouse and rat) appear to be evolving more than twice as fast as primate genomes (humans and chimpanzees). |
|
Gallas gallus (chicken) |
1,000 |
20-23 |
One-third the size of the human genome; genetic variation among domestic chickens seems much higher than in humans. |
|
Pugu rubripes (pufferfish) |
365 |
35 |
The Fugu genome is only one-ninth the size of the human genome, yet it contains 10,000 more genes. |
Invertebrates |
||||
|
Caenorhabditis elegans (nematode) |
97 |
21 |
The fact that every cell of C. elegans has been identified makes its genome a particularly powerful tool in developmental biology. |
|
Drosophila melanogaster (fruit fly) |
137 |
13 |
Drosophila telomere regions lack the simple repeated segments that are characteristic of most eukaryotic telomeres. About one-third of the genome consists of gene-poor centric heterochromatin. |
|
Anopheles gambiae (mosquito) |
278 |
15 |
The extent of similarity between Anopheles and Drosophila is approximately equal to that between human and pufferfish. |
|
Nematostella vectensis (sea anemone) |
450 |
18 |
The genome of this cnidarian is much more like vertebrate genomes than nematode or insect genomes that appear to have become streamlined by evolution. |
Plants |
||||
|
Oryza sativa (rice) |
430 |
33-50 |
The rice genome contains only 13% as much DNA as the human genome, but roughly twice as many genes; like the human genome, it is rich in repetitive DNA. |
|
Populus trichocarpa (cottonwood tree) |
500 |
45 |
This fast-growing tree is widely used by the timber and paper industries. Its genome, fifty times smaller than the pine genome, is one-third heterochromatin. |
Fungi |
||||
|
Saccharomyces cerevisiae (brewer's yeast) |
13 |
6 |
S. cerevisiae was the first eukaryotic cell to have its genome fully sequenced. |
Protists |
||||
|
Plasmodium falciparum (malaria parasite) |
23 |
5 |
The Plasmodium genome has an unusually high proportion of adenine and thymine. Scarcely 5,000 genes contain the bare essentials of the eukaryotic cell. |
Sequencing DNA
In sequencing DNA, the DNA of unknown sequence is first cut into fragments. Each DNA fragment is then copied (amplified), so there are thousands of copies of the fragment. The DNA fragments are then mixed with copies of DNA polymerase, copies of a primer (recall from chapter 11 that DNA polymerase can only add nucleotides onto an existing strand of nucleotides), a supply of the four nucleotide bases, and a supply of four different chain-terminating chemical tags. The chemical tags act as one of the four nucleotide bases in DNA synthesis, undergoing complementary base pairing. First, heat is applied to denature the double-stranded DNA fragments. The solution is then allowed to cool, allowing the primer (the lighter blue box in figure 13.1 1) to bind to a single strand of the DNA, and synthesis of the complementary strand proceeds. Whenever a chemical tag is added instead of a nucleotide base, the synthesis stops, as shown in the figure. For example, the terminating red “T” was added after three normal nucleotides and synthesis stopped. Because of the relatively low concentration of the chemical tags compared with the nucleotides, a tag that binds to G on the DNA fragment, for example, will not necessarily be added to the first G site. Thus, the mixture will contain a series of double-stranded DNA fragments of different lengths, corresponding to the different distances the polymerase traveled from the primer before a chain-terminating tag was incorporated (six are shown in 1).
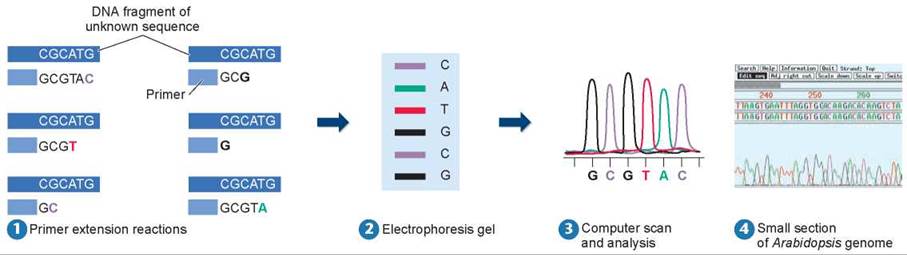
Figure 13.1. How to sequence DNA.
1 DNA is sequenced by adding complementary bases to a single-stranded fragment. DNA synthesis stops when a chemical tag is inserted instead of a nucleotide, resulting in different sizes of DNA fragments. 2 The DNA fragments of varying lengths are separated by gel electrophoresis, the smaller fragments migrating farther down the gel. (The boldface letters indicate the chemical tags added in step 1 that stopped the replication process.) 3 Computers scan the gel, from smallest to largest fragments, and display the DNA sequence as a series of colored peaks. 4 Data from an automated DNA-sequencing run show the nucleotide sequence for a small section of the Arabidopsis (plant) genome.
The series of fragments are then separated according to size by gel electrophoresis. The fragments become arrayed like the rungs of a ladder, each rung being one base longer than the one below it. Compare the lengths of the fragments in 1 and their positions on the gel in 2. The shortest fragment has only one nucleotide (G) added to the primer, so it is the lowest rung on the gel. In automated DNA sequencing, fluorescently colored chemical tags are used to label the fragments, one color for each type of nucleotide. Computers read the colors on the gel to determine the DNA sequence and display this sequence as a series of colored peaks (3 and 4). The development of automated sequencers in the mid-1990s has allowed the sequencing of large eukaryotic genomes. A research institute with several hundred such instruments can sequence 100 million base pairs every day, with only 15 minutes of human attention!
Key Learning Outcome 13.1. Powerful automated DNA sequencing technology is now revealing the DNA sequences of entire genomes.