THE LIVING WORLD
Unit Three. The Continuity of Life
13.3. A Scientific Revolution
In recent years, genetic engineering—the ability to manipulate genes and move them from one organism to another— has led to great advances in medicine and agriculture (figure 13.4). Most of the insulin used to treat diabetes is now obtained from bacteria that contain a human insulin gene. In late 1990, the first transfers of genes from one human to another were carried out to correct the effects of defective genes in a rare genetic immune disorder called Severe Combined Immunodeficiency (SCID)—often called the “Bubble Boy Disorder” based on a young boy who lived out his life in an enclosed, germ-free environment. In addition, cultivated plants and animals can be genetically engineered to resist pests, grow bigger, or grow faster.
The first stage in any genetic engineering experiment is to chop up the “source” DNA to get a copy of the gene you wish to transfer. This first stage is the key to successful transfer of the gene, and learning how to do it is what has led to the genetic revolution. The trick is in how the DNA molecules are cut. The cutting must be done in such a way that the resulting DNA fragments have “sticky ends” that can later be joined with another molecule of DNA.
This special form of molecular surgery is carried out by restriction enzymes, also called restriction endonucleases, which are special enzymes that bind to specific short sequences (typically four to six nucleotides long) on the DNA. These sequences are very unusual in that they are symmetrical— the two strands of the DNA duplex have the same nucleotide sequence, running in opposite directions! The sequence in figure 13.5, for example, is GAATTC. Try writing down the sequence of the opposite strand: it is CTTAAG—the same sequence, written backward. This sequence is recognized by the restriction enzyme EcoRI. Other restriction enzymes recognize other sequences.
What makes the DNA fragments “sticky” is that most restriction enzymes do not make their incision in the center of the sequence; rather, the cut is made to one side. In the sequence in figure 13.5 1, the cut is made on both strands between the G and A nucleotides, G/AATTC. This produces a break, with short, single strands of DNA dangling from each end. Because the two single-stranded ends are complementary in sequence, they could pair up and heal the break, with the aid of a sealing enzyme—or they could pair with any other DNA fragment cut by the same enzyme because all would have the same single-stranded sticky ends. Figure 13.5 2 shows how DNA from another source (the orange DNA) also cut with EcoRI has the same sticky ends as the original source DNA. Any gene in any organism cut by the enzyme that attacks GAATTC sequences will have the same sticky ends, and can be joined to any other with the aid of a sealing enzyme called DNA ligase 3.
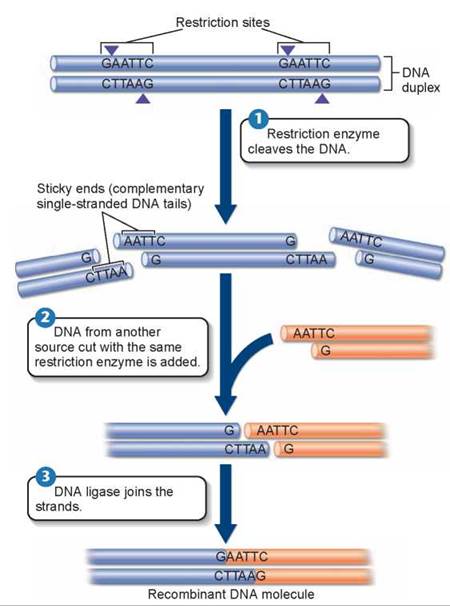
Figure 13.5. How restriction enzymes produce DNA fragments with sticky ends.
The restriction enzyme EcoRI always cleaves the sequence GAATTC between G and A. Because the same sequence occurs on both strands, both are cut. However, the two sequences run in opposite directions on the two strands. As a result, single-stranded tails are produced that are complementary to each other, or "sticky."
Formation of cDNA
As already mentioned, eukaryotic genes are encoded in segments called exons separated from one another by numerous nontranslated sequences called introns. The entire gene is transcribed by RNA polymerase, producing what is called the primary RNA transcript (figure 13.6). Before a eukaryotic gene can be translated into a protein, the introns must be cut out of this primary transcript. The fragments that remain are then spliced together to form the mRNA, which is eventually translated in the cytoplasm. When transferring eukaryotic genes into bacteria (discussed in section 13.4), it is necessary to transfer DNA that has had the intron information removed because bacteria lack the enzymes to carry out this processing. Bacterial genes do not contain introns. To produce eukaryotic DNA without introns, genetic engineers first isolate from the cytoplasm the processed mRNA corresponding to a particular gene. The cytoplasmic mRNA has only exons, properly spliced together. An enzyme called reverse transcriptase is then used to make a complementary DNA strand of the mRNA. A double-stranded DNA molecule is then produced. Such a version of a gene is called complementary DNA, or cDNA.
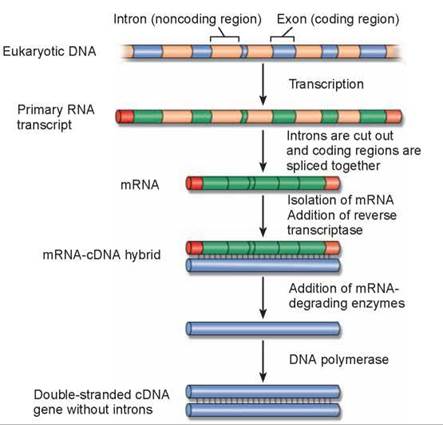
Figure 13.6. cDNA: Producing an intron-free version of a eukaryotic gene for genetic engineering.
In eukaryotic cells, a primary RNA transcript is processed into the mRNA. The mRNA is isolated and converted into cDNA.
cDNA technology is being used in other ways, such as determining the patterns of gene expression in different cells. Every cell in an organism contains the same DNA, but genes are selectively turned on and off in any given cell. Researchers can see what genes are actively expressed using cDNAs.
DNA Fingerprinting and Forensic Science
DNA fingerprinting is a process used to compare samples of DNA. Just as fingerprinting revolutionized forensic evidence in the early 1900s, so DNA fingerprinting is revolutionizing it today. A hair, a minute speck of blood, a drop of semen, all can serve as sources of DNA to convict or clear a suspect.
The process of DNA fingerprinting uses probes to fish out particular sequences to be compared from the thousands of other sequences in the human genome. Because different people have different DNA sequences throughout their genome, they will have different restriction enzyme cutting sites, and so produce different-sized fragments that move to different locations on a gel. Radioactive probes bind to particular locations that occur randomly many times in the genome, “lighting up” the fragments that contain that sequence. If several different probes are used, the chance of any two individuals having the same gel pattern is less than one in a billion. DNA fragments that bind the probes are then visible on autoradiographic film as dark bands. The autoradiograph gel patterns are in essence DNA “fingerprints” that can be used in criminal investigations and other identification applications.
In the first time DNA evidence was used in a court of law, DNA probes were used to characterize DNA isolated from a rape victim’s blood, from semen left by the rapist, and from the suspect’s blood:

You can see that the suspect’s pattern (indicated in pink) matches that of the rapist, and is not at all like that of the victim. Other probes produced similar results. On November 6, 1987, the jury returned a verdict of guilty, the first time a person in the United States was convicted of a crime based on DNA evidence. Since this verdict, DNA fingerprinting has been admitted as evidence in thousands of court cases.
PCR Amplification
Tiny DNA samples, as little as that found in a single human hair, can be magnified to millions of copies by a process called PCR (for polymerase chain reaction). The use of DNA in forensic analysis often depends critically on this PCR technology, developed in 1983 by Kary Mullis. In PCR, a doublestranded DNA fragment is heated so it becomes single stranded; each of the strands is then copied by DNA polymerase to produce two double-stranded fragments. The fragments are heated again and copied again to produce four doublestranded fragments. This cycle is repeated many times, each time doubling the number of copies, until enough copies of the DNA fragment exist for analysis (figure 13.7).
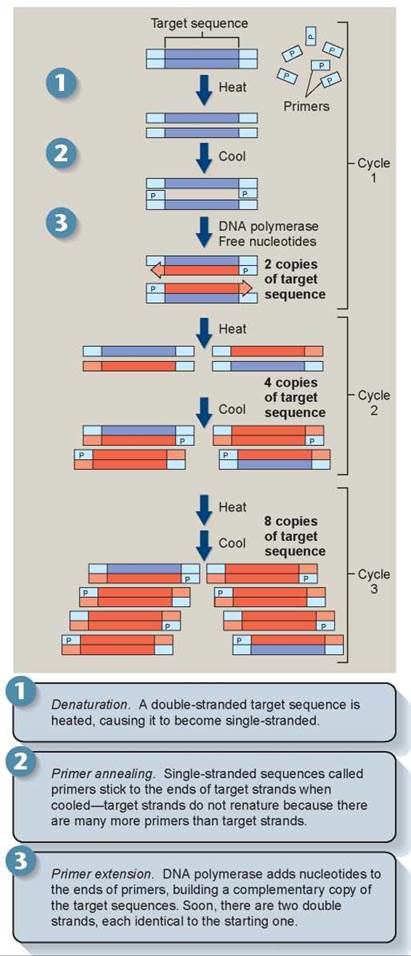
Figure 13.7. How the polymerase chain reaction works.
The amount of DNA found in a single hair will do. Biologists used to think that DNA was present only in the cells at the root of a hair, not in the hair shaft made of the protein keratin. But we now know follicle cells become incorporated into the growing shaft and their DNA is sealed in by the keratin, protecting it from being degraded by bacteria and fungi.
Key Learning Outcome 13.3. Restriction enzymes, the key tools that make genetic engineering possible, bind to specific short sequences of DNA and cut the DNA. This produces fragments with "sticky ends,” which can be rejoined in different combinations.
Today’s Biology
DNA and the Innocence Project
Every person's DNA is uniquely their own, a sequence of nucleotides found in no other person. Like a molecular social security number a billion digits long, the nucleotide sequence of an individual's genes can provide proof-positive identification of a rapist or murderer from DNA left at the scene of a crime—proof more reliable than fingerprints, more reliable than an eye witness, even more reliable than a confession by the suspect.
Never was this demonstrated more clearly than on May 16, 2006 in a Rochester, New York courtroom. There a judge freed convicted murderer Douglas Warney after 10 years in prison.
The murder occurred on New Year's Day in 1996. The bloody body of one William Beason, a prominent community activist, was found in his bed. Police called in all the usual suspects, in this case everyone known to be an acquaintance of the victim. Warney, an unemployed 34-year-old who had dropped out of school in the eighth grade, committed robberies, and worked as a male hustler, learned that detectives wanted to speak to him about the killing, and went to the police station for questioning. Within hours he was charged with murder.
Warney's interrogation was not recorded, but it resulted in a signed confession based on the words that the detective sergeant said Warney uttered, a confession that contained accurate details about the murder scene that had not been made public. Warney said that the victim was wearing a nightgown and had been cooking chicken in a pot, and that the murderer had used a 12-inch serrated knife. The case against him in court rested almost entirely on these vivid details. Even though Warney recanted his confession at his trial, the accuracy of his confession was damning. The details that Warney provided to the sergeant could have come only from someone who was present at the crime scene.
There were problems with his confession, brought out at trial. Three elements of the signed statement's account of that night were clearly not true. It said Warney had driven to the victim's house in his brother's car, but his brother did not own a car; it said Warney disposed of his bloody clothes after the stabbing in the garbage can behind the house, but the can, buried in snow from the day of the crime, did not contain bloody clothes; it named a relative of Warney as an accomplice, but that relative was in a secure rehabilitation center on that day.
The most difficult bit of evidence to match to the written confession was blood found at the scene that was not that of the victim. Drops of a second person's blood were found on the floor and on a towel. The difficult bit was that the blood was a different blood type than Warney's.
At trial, prosecutors pointed out that the blood could have come from the accomplice mentioned in the confession, and pounded away on the point that the details in the confession could only have come from firsthand knowledge of the crime.
After a short trial, Douglas Warney was convicted of the murder of William Beason and sentenced to 25 years.
When appeals failed, Mr. Warney in 2004 sought help from the Innocence Project, a nonprofit legal clinic that helps identify wrongly convicted individuals and secure their freedom. Set up at the Benjamin N. Cardozo School of Law in New York by lawyers Barry Scheck and Peter Neufeld, the Innocence Project specializes in using DNA technology to establish innocence.
The Innocence Project staff petitioned the court for additional DNA testing using new sensitive DNA probes, arguing that this might disclose evidence that would have resulted in a different verdict. The judge refused, ruling that the possibility that the blood found at the scene of the crime might match that of a criminal already in the state databank was "too speculative and improbable” to warrant the new tests.
Someone in the prosecutor's office must have been persuaded, however. Without notifying Warney's legal team, Project Innocence, or the court, this good Samaritan arranged for new DNA tests on the blood drops found at the crime scene. When compared to the New York State criminal DNA database, they hit a strong match. The blood was that of Eldred Johnson, in prison for slitting his landlady's throat in Utica two weeks before Beason was killed.
When confronted, the prisoner Johnson readily admitted to the stabbing of Beason. He said he was the sole killer, and had never met Douglas Warney.
This leaves the interesting question of how Warney's signed confession came to include such accurate information about the crime scene. It now appears he may have been fed critical details about the crime scene by the homicide detective leading the investigation, who has since died.
DNA testing has become a pillar of the American criminal justice system. It has provided key evidence that has established the guilt of thousands of suspects beyond any reasonable doubt—and, as you see here, also provided the evidence that our criminal justice system sometimes convicts and sentences innocent people. Over more than a decade, the Innocence Project and other similar efforts have cleared hundreds of convicted people, strong proof that wrongful convictions are not isolated or rare events. DNA testing opens a window of hope for the wrongly convicted.