SAT Biology E/M Subject Test
Part II: Subject Review
Chapter 10 Microorganisms
There are four types of microorganisms you should know about for the SAT Biology E/M Subject Test: protists, fungi, bacteria, and viruses. Because we”ve already covered everything you need to know about protists in Chapter 9, in this chapter we”ll cover fungi, bacteria, and viruses.
LET”S TALK ABOUT FUNGI
Most fungi are multicellular eukaryotes. That means they are made up of many cells, and that their cells have nuclei and other organelles. In some fungi, the cells are not separated by cell walls (fungal cell walls, by the way, are made of chitin), and they seem to be one giant cell with many nuclei—in other words, multinucleate. In other fungi, the cells are kept distinct. There are many examples of fungi that you are familiar with—molds, mushrooms, etc.
Something to Remember
About Yeast
Often people want to
classify yeast as
prokaryotic because
yeasts are unicellular, but
don”t fall into that trap.
Yeasts are fungi, and all
fungi are eukaryotes—
they have nuclei and organelles.
Fungi lack chloroplasts, which means they cannot photosynthesize (produce their own food). Therefore, they are classified as heterotrophs. Furthermore, they are absorptive feeders. This means they secrete hydrolytic enzymes that digest their food outside their bodies. This “predigested” food can then be easily absorbed by the fungus. Many fungi feed on dead or decaying material, helping to break it down; they are decomposers.
Fungi reproduce in several different ways:
1. Asexual spores: These spores are kind of like seeds that can drop off the fungus and grow a new organism.
2. Sexual spores: Fungi can produce sexual spores (kind of like sperm and ova) that combine to form a new organism.
3. Vegetative growth: In this type of reproduction, a portion of the fungus breaks off and forms a new fungus.
4. Budding: A new fungus grows off the side of the old fungus. An example of a fungus that reproduces in this way is yeast.
LET”S TALK ABOUT BACTERIA
Bacteria reproduce asexually in a process called binary fission. It”s very simple—the bacterium replicates its single chromosome, then splits in half, creating two new genetically identical bacteria. Bacteria are single-celled organisms, and these organisms are the only prokaryotes around. Prokaryotes have no nuclei and no membrane-bound organelles (they do have ribosomes, which are not membrane-bound). Because they have no nuclei, their DNA is found in the cytoplasm. Bacterial DNA is usually found as a single, circular chromosome. It”s still double-stranded, but the ends of the strand are joined in a circle. This helps protect the DNA from damage. Bacteria also have a cell wall, which is made of peptidoglycan (proteins and sugars).
Bacteria reproduce in a process called binary fission. It”s very simple—the bacterium replicates its single chromosome, then splits in half. Each of the new daughter cells gets one of the chromosomes and about half the organelles and cytoplasm. This is a great way of increasing the numbers of bacteria—one becomes two, which become four, which become eight, and so on—but this does not allow for any type of genetic recombination. In other words, there is no chance of mixing up the DNA using this type of reproduction. As we”ve seen in Chapter 9, being able to mix up the DNA a bit gives a species variability, which can lead to evolution, so it”s definitely an advantage to be able to do this.
Bacteria can achieve genetic recombination—mix up the DNA—in three different ways:
1. Transformation: In transformation, bacteria can pick up new DNA from the extracellular environment. This is unusual and relatively rare in natural environments but is often used in research for a number of different purposes.
2. Conjugation: Conjugation means that a bacterium replicates its DNA and donates some of it to another bacterium through a bridge called a pilus.
3. Transduction: In transduction, a virus carries DNA from one bacterium to another bacterium during infection.
How This Information Might Show Up on the Test
Bacterial genetics often appears on the SAT Biology E/M Subject Test in an experiment-type question in which one strain of bacteria is being grown, then something happens to it, and suddenly it displays different properties than it did before—it becomes a new strain. For example, perhaps two different strains of bacteria (Strain A and Strain B) were being grown separately, then they were mixed, and a third, new strain (Strain C) resulted. Strain A is resistant to the antibiotic ampicillin (it is not killed by ampicillin), but sensitive to tetracycline (tetracycline kills it). Strain A would be denoted ampr tets. Strain B is sensitive to ampicillin, but resistant to tetracycline, so it would be denoted amps tetr. The new strain, Strain C, is resistant to both antibiotics, so it”s denoted ampr tetr.
Antibiotic resistance is determined by the bacterium”s DNA, so if the resistance or sensitivity to an antibiotic changes, something has changed in the bacterium”s DNA—in other words, genetic recombination has occurred. But how do you know which type of recombination? Was ittransformation? Conjugation? Transduction? Here”s a handy table to help you figure it out.
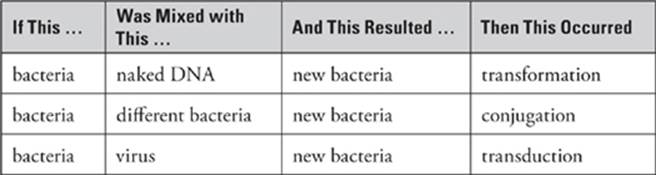
So in our example above, Strain A was mixed with Strain B to produce Strain C. Because bacteria were mixed with different bacteria to produce new bacteria, conjugation must have occurred. (More specifically, Strain B must have donated the DNA for tetracycline resistance to Strain A, resulting in Strain C.)
How Bacteria Get Nutrition
Some bacteria, the cyanobacteria, can perform photosynthesis, which means that they”re autotrophs. Most bacteria, however, cannot perform photosynthesis, and they”re heterotrophs. The heterotrophs get their nutrition from other organisms.
Bacteria
Saprobes are bacteria
that are decomposers and
obtain their nutrients from
the breakdown of dead
organic matter.
Parasites get their
nutrition from living hosts,
harming the host
in the process.
Symbionts also get their
nutrition from living hosts,
but they do not harm the
host in the process.
Most bacteria need oxygen. They”re obliged to have oxygen, just as we are, and they are known as obligate aerobes. Some bacteria are poisoned by oxygen, and these are known as obligate anaerobes. Finally, there are some bacteria that will use oxygen if it”s available but can survive by fermentation if it”s not. These bacteria are facultative anaerobes. Here”s a quick summary table for oxygen use by bacteria.

Let”s Talk About Auxotrophs
Remember from the discussion on plants that the suffix troph refers to nutrition. The prefix auxo is derived from the word auxiliary, which means “supplementary.” So an auxotroph is an organism that requires supplementary nutrition.
Most bacteria are not auxotrophs and are referred to as wild type. These bacteria are perfectly capable of synthesizing all the building blocks they need in terms of nutrition, as long as they are provided with a carbon source such as glucose. They can make all the amino acids, lipids, etc., that they need to build their larger macromolecules (peptidoglycan, carbohydrates, proteins, etc.). For researchers in a laboratory, these bacteria are simple to grow. Wild types grow easily in a petri dish on agar plates with glucose, or in a liquid medium with glucose in a test tube.
Auxotrophic bacteria, however, need a little help. They need additional substances put into their growth medium along with the glucose. What additional substances? That depends on the type of auxotroph you”re dealing with. Most auxotrophic bacteria lack the ability to synthesize some amino acid. They are denoted by the amino acid they cannot make, along with a minus sign. For example, if a bacterium could not synthesize arginine, it would be an arg– auxotroph. If a different bacterium could not synthesize leucine, it would be a leu– auxotroph.
A single strain of bacteria might have a combination of traits. For example, it might be unable to synthesize leucine but able to synthesize arginine. This strain would be denoted leu– arg+. Perhaps it is also unable to synthesize histidine. If that were the case, it would be denoted leu– arg+his–. And so on. Here”s an important point to remember:
If the substance that cannot be synthesized is not added to the growth medium, the auxotroph cannot grow.
However, other bacteria that are not auxotrophic for that particular substance can grow just fine.
Lactose Intolerant?
Wildtype bacteria can
grow on pretty much
any carbon source, like
glucose or lactose. Some
bacteria, denoted lac-, are
unable to utilize lactose
and can only be grown on
glucose.
How This Might Show Up on the Test
This most often appears as a question in which you are asked to determine the characteristics of a strain of bacteria. A researcher attempts to grow a strain of bacteria in several different media, each containing or lacking some amino acid. Sometimes the bacteria grow and sometimes they don”t. You have to figure out if the strain is auxotrophic and what type of auxotroph it is. Here is a simple two-step technique to figure it out:
Step 1: Determine which amino acid is missing from the medium.
Step 2: See if the bacteria grow there.
YES: The bacteria are not auxotrophic for that amino acid (denoted with a plus sign).
NO: The bacteria are auxotrophic for that amino acid (denoted with a minus sign).
Note that you will have to do this separately for each situation.
Let”s Put It Into Practice
The table below contains data from an experiment in which the researcher attempted to grow three different strains of bacteria (Strain A, Strain B, and Strain C) in various types of growth media. Analyze the data, and see if you can figure out the characteristics of each strain.
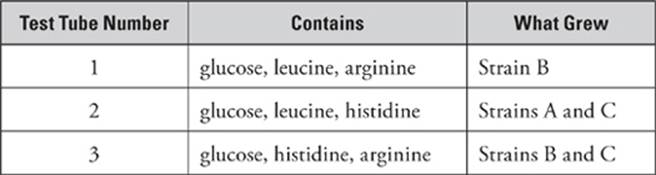
In this problem, there are three different test tubes and three different strains of bacteria. So you must do Step 1 three times, once for each tube, and Step 2 three times, once for each strain. The first step is to figure out which amino acid is missing. Note that you need to concern yourself only with the amino acids being used—you don”t have to worry about all 20! In this experiment, only leucine, arginine, and histidine are being considered.
It”s in the Genes
The ability to synthesize
an amino acid is coded in
the DNA. In other words,
this is a genetic trait.
These traits might show
up in genetic
recombination
experiments and
questions, like the
previous example about
antibiotic resistance and
conjugation.
Tube 1
Step 1: Missing histidine
Step 2: Strain A did not grow. Strain A is his–. Strain B did grow, so Strain B is his+. Strain C did not grow, so Strain C is his–.
Tube 2
Step 1: Missing arginine
Step 2: Strain A did grow. Strain A is arg+. Strain B did not grow, so Strain B is arg–. Strain C did grow, so Strain C is arg+.
Tube 3
Step 1: Missing leucine
Step 2: Strain A did not grow. Strain A is leu–. Strain B did grow, so Strain B is leu+. Strain C did grow, so Strain C is leu+.
So the final summary is:
Strain A: his– arg+ leu–
Strain B: his+ arg– leu+
Strain C: his– arg+ leu+
Remember that if a bacterium can grow in the absence of a particular amino acid, it is not auxotrophic for that amino acid.
Another Important Thing About Bacteria—They Supply Nitrogen to Soil
Nitrogen is an important component of protein, and plants need nitrogen to grow properly. The atmosphere is about 80 percent nitrogen, but plants cannot use gaseous nitrogen (N2); they must have it in the form of nitrate (NO3–) or ammonium (NH4+).
Much of the soil nitrogen comes from the breakdown of dead organic material that releases nitrogen. This nitrogen is converted to ammonium by ammonifying bacteria. Some of the soil nitrogen comes from gaseous nitrogen in the atmosphere, which is converted to ammonia by nitrogen-fixing bacteria. The ammonia is converted to ammonium in the soil. Finally, the ammonium is converted to nitrate by yet another type of bacteria, the nitrifying bacteria.
For the SAT Biology E/M Subject Test, the most important thing to remember about nitrogen and bacteria is that some nitrogen-fixing bacteria form a symbiotic relationship with the roots of certain plants. This relationship is mutualistic, meaning that both the plant and the bacteria benefit from the association. The bacteria live in the roots of these plants, in structures called nodules. They supply the plant with fixed nitrogen, and the plant supplies them with nutrients. Plants that establish this type of mutualistic relationship with nitrogen-fixing bacteria are legumes—beans, peas, peanuts, clover, alfalfa, and soybeans. Sometimes the bacteria fix so much nitrogen that the plant can”t use it all, and the extra is secreted into the soil. This is one reason for crop rotation—a legume is planted in the field first and harvested, then a non-legume is planted, which can use the “leftover” nitrogen from the previous legume crop.
LET”S TALK ABOUT VIRUSES
A virus isn”t really considered a cell, and it”s not even technically considered to be alive. It has only two components: (1) a coat made of protein (only protein) called the capsid and, inside the coat, (2) nucleic acid (the genome). Depending on the virus, the genome might be DNA or RNA.
Because viruses consist only of a protein capsid and a nucleic acid genome, they have none of the machinery required to reproduce themselves. Therefore, they can reproduce only with the help of another cell, using that cell”s enzymes as needed. All types of cells can be infected by viruses—bacteria, fungi, plants, or animals. Let”s consider a virus with a DNA genome.
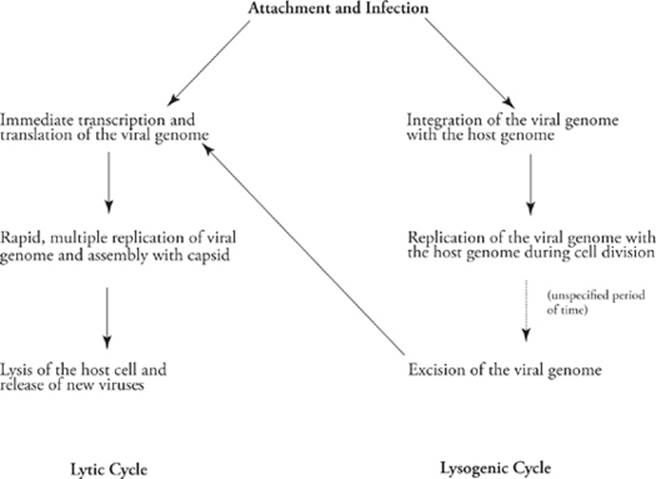
The viral life cycle begins with the following two steps:
1. Attachment: The virus attaches itself to the host cell. This is a specific interaction between the virus and the host cell; a virus can attach only to its particular host.
2. Infection: The virus injects its genome into the host cell.
Once inside the host, the viral genome enters into one of two possible life cycles, the lytic cycle or the lysogenic cycle. In the lytic cycle, the viral genome is first transcribed and translated (using the host”s RNA polymerases and ribosomes) to make viral proteins (for example, the capsid proteins). Host DNA polymerases then replicate the viral genome, and the new viral genomes are packaged into the new viral capsids. Finally, the host is broken open (lysed) by a special viral enzyme and the new viruses escape to infect new hosts. Because viruses absolutely cannotreproduce outside a host cell, and because the host cell is harmed in the process, viruses are considered to be parasites.
In the lysogenic cycle, the viral genome is not immediately transcribed, translated, and replicated. Instead, it is inserted (integrated) into the host”s genome, where it remains dormant. Every time the host replicates its genome (during cell division) the viral genome is replicated, too. Thus, every daughter cell of the original infected host is also infected; they carry the viral genome integrated with their own. The virus can remain in this dormant state for a long time—years and years in some cases. If the host cell experiences stress of some sort (illness, for example) the virus can remove itself from the host genome and begin the lytic cycle, being transcribed, translated, etc. An example of a virus that follows the lysogenic cycle is the human virus that causes cold sores (HSV 1).
Here”s a side-by-side comparison of the two cycles:
A Special Note About Animal Viruses
Because animal cells do not have a cell wall, viruses that infect these cells do not necessarily have to lyse them to escape. The new viral particles can simply bud out through the plasma membrane. This is an advantage to the virus, because the cell stays alive longer, and ultimately more viruses can be produced (lysing a cell kills it instantly, and if the cell dies, viral production stops). As the virus buds out, it becomes coated in plasma membrane; this layer of membrane around the capsid is called an envelope.
Viruses with RNA Genomes
Viruses with RNA genomes have a slight problem when it comes to replicating their genome. Every other organism on the planet has a DNA genome; so they have the DNA polymerases necessary for replication. If a virus also has a DNA genome, it has no problem “borrowing” the host cell”s DNA polymerase to replicate itself (recall that viruses rely on host cell enzymes for transcription, translation, and replication).
But an RNA virus cannot use DNA polymerase to replicate (DNA polymerase copies only DNA); the virus needs an RNA polymerase. Furthermore, the virus can”t use the host”s RNA polymerase, because that enzyme makes RNA only by reading a DNA template (transcription). The virus needs a special enzyme that makes a strand of RNA by reading an RNA template—an RNA-dependent RNA polymerase. Because this is a unique viral-only enzyme, the virus must either (a) carry the enzyme with it in its capsid and inject it into the host along with its genome or (b) synthesize the protein during translation of the viral genome (a part of the normal viral life cycle).
Retroviruses
Retroviruses are RNA viruses that go through the lysogenic life cycle. Recall that in this life cycle, the viral genome is integrated with the host genome. Because in this case the viral genome is RNA, and all other organisms have a DNA genome, a DNA copy of the viral genome must first be made before integration can occur. Again, this requires a special enzyme, because the host has no enzymes that make DNA by reading an RNA template. This enzyme is an RNA-dependent DNA-polymerase, and, as with the RNA-dependent RNA-polymerase discussed above, must either be carried with the virus or synthesized during translation. The common name for RNA-dependent DNA-polymerase is reverse transcriptase—normally transcription creates RNA from DNA, and this enzyme runs reverse transcription; it creates DNA from RNA. The virus that causes AIDS is a retrovirus (HIV), and many cancer-causing viruses are retroviruses.
Quick Quiz #1
Fill in the blanks and check the appropriate boxes:
1. Fungi are classified as [ ![]() eukaryotes
eukaryotes ![]() prokaryotes ].
prokaryotes ].
2. Bacteria [ ![]() do
do ![]() do not ] have mitochondria.
do not ] have mitochondria.
3. Bacteria [ ![]() do
do ![]() do not ] have ribosomes.
do not ] have ribosomes.
4. Bacteria [ ![]() do
do ![]() do not ] have cell walls.
do not ] have cell walls.
5. Yeast are [ ![]() eukaryotes
eukaryotes ![]() prokaryotes ] that reproduce by ______.
prokaryotes ] that reproduce by ______.
6. A viral _________________________ is made of protein.
7. [ ![]() Transformation
Transformation ![]() Transduction ] occurs when a virus transfers some DNA from one bacterium to another.
Transduction ] occurs when a virus transfers some DNA from one bacterium to another.
8. Fungi [ ![]() can
can ![]() cannot ] photosynthesize.
cannot ] photosynthesize.
9. Bacteria that are perfectly happy in the presence OR absence of oxygen are called _________________________________.
10. Number the following steps of the lytic cycle in the order they would occur:
_____ Virus injects nucleic acid into host cell.
_____ Host cell is lysed and new viruses are released.
_____ Viral capsid attaches to host cell.
_____ New capsids are formed.
_____ Viral nucleic acids are replicated using host cell machinery.
11. [ ![]() Some
Some ![]() All ] fungi are eukaryotes.
All ] fungi are eukaryotes.
12. A [ ![]() parasitic
parasitic ![]() mutualistic ] relationship exists between some plants and ______________________________ bacteria. These types of plants are called ____________________.
mutualistic ] relationship exists between some plants and ______________________________ bacteria. These types of plants are called ____________________.
13. When a bacterium replicates its DNA and gives some of the DNA to another bacterium through a pilus, this is called ______________________________.
14. Fungi have a cell wall made of [ ![]() proteoglycan
proteoglycan ![]() chitin ].
chitin ].
15. Bacteria are classified as [ ![]() prokaryotes
prokaryotes ![]() eukaryotes ] and therefore [
eukaryotes ] and therefore [ ![]() do
do ![]() do not ] have nuclei.
do not ] have nuclei.
16. An enzyme that makes a strand of RNA by reading a strand of DNA is called ____________________.
17. Retroviruses go through the [ ![]() lytic
lytic ![]() lysogenic ] life cycle.
lysogenic ] life cycle.
Correct answers can be found in Chapter 15.
COOL STUFF: RECOMBINANT DNA TECHNOLOGY
Now that you know about bacteria, viruses, and DNA, let”s talk about how these microorganisms can be used in molecular biology research. First, we have to cover a little background information.
Restriction Enzymes
Restriction enzymes are enzymes that recognize a particular short DNA sequence and then cut the DNA strand within that sequence. They were first discovered in bacteria. Bacteria use the enzymes to cut (and thereby destroy) foreign DNA, such as viral DNA. This would restrict the growth of the virus, hence the name restriction enzymes. The bacterium”s own DNA is protected in some way, often by the addition of a methyl group (–CH3) to the sequence recognized by the enzyme, thus preventing the enzyme from binding there.
There have been many, many restriction enzymes discovered. They are named for the strain of bacteria in which they were first found; for example, the restriction enzyme EcoRI was found in E. coli. Each restriction enzyme recognizes a different DNA sequence. That sequence is usually apalindrome (it reads the same from both directions); for example, here is the sequence recognized by EcoRI:
![]()
and here is the sequence recognized by PstI (discovered in Providencia stuartii):
![]()
and here is yet another recognized by HpaI (discovered in Hemophilus parainfluenzae):
![]()
Notice that if you read the sequence (from 5” to 3”) in either direction, it is the same.
A “Blunt” Explanation of a “Sticky” Situation
Restriction enzymes cut the DNA sequence in one of two ways, either staggered or blunt. In a staggered cut, short single-stranded tails called sticky ends are left at the cut ends of the DNA strand. EcoRI and PstI both perform a staggered cut. They cut the DNA strand like this:
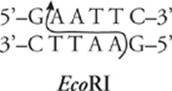
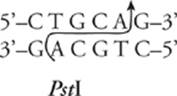
to produce sticky ends like this:
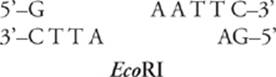
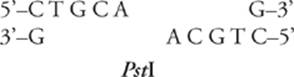
If a piece of DNA is cut with a restriction enzyme that produces sticky ends, it can be reattached—ligated—to any other piece of DNA cut with the same restriction enzyme. The single-stranded tails must overlap in a complementary fashion.
In a blunt cut, the DNA is cut straight across both strands. HpaI performs a blunt cut like this:

to produce blunt ends like this:

If a piece of DNA is cut with an enzyme that produces blunt ends, it can be ligated to any other piece of DNA cut with any other enzyme that produces blunt ends, because the ends do not need to overlap in a complementary fashion.
Plasmids
A plasmid is a small circular piece of DNA frequently found in bacteria and sometimes in yeast. The plasmid is not considered to be part of the bacterial or yeast genome and can reproduce independently of the microorganism. And because the plasmid is DNA, its sequence contains restriction sites. Let”s take a look at how plasmids are used in recombinant DNA research.
Antibiotic Resistance
Some bacteria carry plasmids that contain genes that make the bacteria resistant to antibiotics. These plasmids can be copied, spread throughout the population of bacteria, and even spread to different populations of different bacteria. The result of this process is that many of the bacteria that cause diseases in humans are becoming resistant to certain antibiotics.
Back to Recombinant DNA Technology
Recombinant DNA technology simply refers to the creation of new DNA molecules by cutting and splicing (i.e., recombining) the DNA with restriction enzymes. It is used primarily in the study of DNA sequences of species that do not reproduce rapidly.
Suppose you were a researcher studying a certain mouse gene. As part of your studies, you wish to characterize the DNA for that gene by determining its sequence, or by determining its restriction map (a map of the location of various restriction sites within that piece of DNA). In any case, you”re going to need quite a bit of that DNA. You could sacrifice a bunch of mice, grind them up (eeww!), and extract the DNA from the tissue. This would be a lot of work, and every time you need a sample of DNA you have to repeat this procedure. Further, this would cost a lot for the mice, not to mention the space for breeding the mice, and the cost of feeding the mice, etc. (Plus, it”s hard on the mice!)
But suppose that you obtain a small sample of mouse DNA by sacrificing a single mouse and extracting the DNA from its tissues. And suppose further that you know the particular gene you are interested in is bounded on either side by an EcoRI restriction site. Using EcoRI, you could cut the gene out of its chromosome and separate it from the remainder of the mouse DNA. In reality, there are several steps involved here, but to keep things simple (after all, we just want to get an idea of the big picture), let”s say that these are the only two EcoRI sites in the entire mouse genome. Now you have a small (very small!) sample of just the DNA you are interested in, with an EcoRI sticky end on each side. The next step is to amplify (increase the amount of) that DNA.
So you order a plasmid (custom-made plasmids are commercially available!) that has an EcoRI site in its sequence, and you cut that plasmid with EcoRI. Then you mix the cut plasmid with your DNA sample (that has the EcoRI sticky ends), and because the sticky ends on your DNA are complementary to the sticky ends on the plasmid, they will base-pair with one another and your DNA sample will become inserted into the plasmid. You then add a new enzyme called DNA ligase that permanently joins the newly base-paired regions. You now have a plasmid with your DNA of interest inserted.
Now for the Final Part …
You insert this plasmid into a general strain of laboratory bacteria (okay again, several steps here which, for clarity, will be omitted) and grow gallons of the bacteria. Bacteria are easy and inexpensive to grow, and they reproduce very rapidly, doubling their numbers approximately every 20 minutes. And every time the bacteria reproduce, the plasmid does, too, so you are in effect copying your piece of DNA over and over again. As long as the bacteria are reproducing, you essentially have an endless supply of your DNA of interest. Getting the plasmid out of the bacteria is relatively easy, and removing your DNA of interest is as easy as recutting the plasmid with EcoRI.
Yes, this is an oversimplification of the process, but the general idea is sound. Using recombinant DNA technology, plasmids can be custom-made to reproduce in bacteria or in eukaryotes. Sometimes promoter regions can be added so that DNA inserted after the promoter can be transcribed and translated. This allows the cells containing the plasmid to start making the protein coded for by the DNA, creating an unlimited supply of the protein of interest! Using plasmids and cells (prokaryotic or eukaryotic) to reproduce DNA or make protein is a much more efficient method than using live animals for DNA or protein collection.
Here”s Another Example
Let”s say you have isolated a gene that gives tomato plants the ability to resist attack by certain parasites—perhaps the gene codes for a chemical in the sap of the tomato plant that weakens or kills the parasite. However, the gene is found only in a few isolated tomato plants. If you could somehow get that gene into all the tomato plants on a farm, it would produce a hardier plant and increase crop yield. So, using restriction enzymes, you insert the gene into a lysogenic virus that infects tomato plants, but is relatively harmless (in other words, the virus does not kill the plant or make the plant sick). Then you allow your recombinant virus to infect a field of plants, and as part of its normal life cycle the virus inserts its genome (which now contains the new parasite-resisting gene) into the tomato genome. These new recombinant plants can now resist the effects of the parasite and are minimally affected by the virus.
Here”s another definition: a vector. A vector is simply a shuttle used to move DNA between species. The plasmid was our vector in the first example, and the virus was the vector in the second example.
Remember This
The examples presented here are an oversimplification, but here”s the big picture:
Restriction enzymes give us the ability to effectively “cut and paste” (recombine) DNA into custom combinations.
Using vectors, these custom DNAs can be moved into other organisms. This allows the DNA to be amplified and studied much more rapidly than it could be in mammalian (or even plant) species. Or, it can provide a new gene (or several genes) to an organism that can give it an advantage. This is the basis for gene replacement therapy as treatment for genetic disorders (although this has not yet been successful in humans).
Quick Quiz #2
1. A _________________________ cut of DNA by a restriction enzyme produces sticky ends.
2. A piece of DNA cut with a restriction enzyme that produces sticky ends can be ligated to any piece of DNA cut with [![]() the same restriction enzyme
the same restriction enzyme ![]() any other restriction enzyme that produces sticky ends ].
any other restriction enzyme that produces sticky ends ].
3. The enzyme that seals together cut pieces of DNA is called ________.
4. A sequence of DNA that reads the same from both directions is called a _________________________.
5. [![]() True
True ![]() False ] A piece of DNA cut with a restriction enzyme that produces blunt ends can be ligated into a plasmid cut with any other restriction enzyme.
False ] A piece of DNA cut with a restriction enzyme that produces blunt ends can be ligated into a plasmid cut with any other restriction enzyme.
Correct answers can be found in Chapter 15.
Key Words
fungi
chitin
multinucleate
absorptive feeders
asexual spores
sexual spores
vegetative growth
budding
eukaryotes
prokaryotes
peptidoglycan
binary fission
resistant
sensitive
transformation
conjugation
transduction
obligate aerobes
obligate anaerobes
facultative anaerobes
saprobes
parasites
symbionts
auxotroph
wild type
mutualistic
nodules
legumes
capsid
genome
attachment
infection
lytic cycle
lysed
lysogenic cycle
integrated
envelope
RNA-dependent RNA polymerase
RNA-dependent DNA polymerase
reverse transcriptase
restriction enzymes
EcoRI
sticky ends
blunt cut
plasmid
restriction map
amplify
vector
Summary
• There are four types of microorganisms that you need to know for the Biology E/M test: protists, fungi, bacteria, and viruses.
• Most fungi are multicellular eukaryotes; yeasts are unicellular fungi.
• Fungi can reproduce by asexual spores, sexual spores, vegetative growth, and by budding.
• Bacteria are prokaryotes. Some bacteria are autotrophs and some are heterotrophs.
• Some bacteria are auxotrophs and require help to grow—usually by adding something to their growth medium (like an amino acid).
• Some bacteria supply soil with nitrogen to help plants grow properly.
• Viruses are not cells and are not alive. They can reproduce only with the help of another cell.
• There are two main viral life cycles—the lytic cycle and the lysogenic cycle.
• Recombinant DNA technology allows the creation of custom DNA for study or gene therapy.