CHEMICAL BIOLOGY
Peptidomimetics
Claudio Toniolo and Fernando Formaggio, Institute of Biomolecular Chemistry, Padova Unit, CNR, Department of Chemistry, University of Padova, Padova, Italy
doi: 10.1002/9780470048672.wecb439
This review article provides an introduction to folded and extended conformational motifs of synthetic peptidomimetics as templates for chemical biology applications. Several variants of these 3-dimensional structures have been assessed, which comprise versatile scaffolds for presentation of side-chain and main-chain peptide groups for molecular recognition. Specifically, relevant parameters and stereochemical consequences are discussed for helices of peptidomimetics based on: 1) α-amino acids alkylated (e.g., methylated or ethylated) at the Cα-atom; 2) α,β-didehydroamino acids, in particular α,β-didehydrophenylalanine, with a carbon-carbon double bond between the α- and β-positions; 3) N-alkylated α-amino acids, whereby the side chains typical of coded residues are transferred from the Cα- to the N-atom; 4) β-amino acids, the subclass of residues investigated most extensively in which the amino and carboxyl functionalities are separated by more than one carbon atom; and 5) short-range, backbone-to-backbone cyclizations that generate small-ring, monolactam or dilactam building blocks.
Introduction
Peptidomimetic is a molecule that bears identifiable resemblance to a peptide that, as a ligand of a biologic receptor, can imitate or inhibit the effect of a natural peptide. Numerous peptidomimetics have been developed over the past 25 years by bioorganic and medicinal chemists. Some molecules (i.e., those with modifications in the backbone amide group) also have been termed pseudopeptides or peptide bond surrogates (1). These synthetic analogs of peptides have a variety of applications, but most expanded interest in this area focuses on their potential for developing metabolically stabilized and/or more potent and selective bioactive compounds. The complex synthetic aspects (2) and relevant biological data (1, 3, 4) of peptidomimetics have been summarized extensively in review articles and in book chapters. In this introductory review, we focus on the 3-dimensional (3-D) structural properties of peptidomimetics characterized heavily by a few, well-studied amino acids with modifications in the main chain and/or in the side chain. An effort was made through appropriate citations to expose the nonexpert and the beginning graduate students to the remarkable advances made in this area of research. The field of peptidomimetic chemistry is entering new decades of exploding activity. This time is exciting as new techniques and methodologies have now made it possible to answer questions that were not possible to address even few years ago. Many developments ushered in the new era of peptidomimetic chemistry, which include advancements in solution and in solid-phase peptide synthesis, organic synthesis, nuclear magnetic resonance and related biophysical techniques, and mass spectrometry. These advancements take advantage of an increasing understanding of protein folding. Numerous examples now exist of low molecular weight peptides and peptidomimetics that adopt folded or extended structures, and in some cases they serve as exceptional substitutes for their much larger protein counterparts in life processes. Chemists in both academia and industry are making important strides to push the frontiers in these areas. With the ability to incorporate unnatural amino acids that have strong conformational or nucleating bias, it will be possible to force much smaller peptides and peptidomimetics to adopt appropriate 3-D structures and functions. The future for the young scientists who ultimately want to pursue this line of research is indeed very promising. Rather than looking at peptidomimetics as a mature field, it should be argued that we are only in our infancy in terms of sophistication. Every reason exists to believe that a combination of approaches, which include biophysical and functional characterizations, will propel this area forward in the next years. It is especially important for those who are considering future research areas to analyze seriously the potential of such studies.
Among the various types of known ordered secondary structures adopted by peptides formed exclusively by α-amino acid building blocks, in this review article we shall discuss the classic α-helix, the 310-helix, the 2.05-helix, and the type-I poly(Pro)n helix. Relevant parameters for these helices are given in Table 1.
Table 1. Relevant average parameters for peptide helices based on L-configurated α-amino acids discussed in this review article
|
Parameter |
α-Helix |
310-Helix |
2.05-Helix |
Poly(Pro)n I* |
|
φ (°)1 |
-63 |
-57 |
180 |
-70 |
|
ψ (°)2 |
-42 |
-30 |
180 |
160 |
|
n3 |
3.63 |
3.24 |
2.00 |
-3.30 |
|
d (A)4 |
1.56 |
1.94 |
3.70 |
2.22 |
|
p (A)5 |
5.67 |
6.29 |
7.40 |
7.33 |
*In the poly(Pro)n I helix, all ω(Cia-C'i-Ni+1-Ci+1a) torsion angles are in the unusual cis conformation (0°).
1The (C'i-1-Ni-Cai-C'i) torsion angle.
2The (Ni-Cai-C'i-Ni+1) torsion angle.
3Number of amino acids per helical turn (positive values refer to right-handed helices, whereas negative values refer to left-handed helices)
4Axial translation (per residue).
5Pitch or axial translation per helical turn.
Peptides Based on Ca-Alkylated α-Amino Acids
Peptides rich in the noncoded Ca-methylated α-amino acids (1) are biased strongly to fold tightly into the intramolecularly H-bonded, α-helical, or the strictly related 310-helical conformation (5-9), the latter generated by sequences of helical β-turns (10, 11). The same 3-D structural propensities are shared by the Ca,a-cyclized α-amino acids (2).
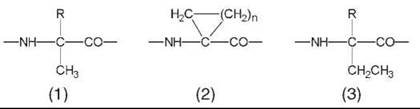
By inserting appropriate amino acid side chains (e.g., hydrophilic and hydrophobic), it is feasible to construct amphiphilic helices (with two faces that possess different properties). The conformational strategy should take into account the parameters typical of each of the two helices, in particular that: 1) The α-helix (Fig. 1a) is characterized by a fractional number of amino acids per turn (![]() 3.5) and consequently its smallest repeat (i.e., the shortest main-chain length that brings two side chains exactly one on top of the other) is a heptad (7 residues); and 2) in the 310-helix (Fig. 1b), which has an integer number of amino acids per turn (≅3.0), a triplet of residues selected carefully will produce the expected amphiphilicity.
3.5) and consequently its smallest repeat (i.e., the shortest main-chain length that brings two side chains exactly one on top of the other) is a heptad (7 residues); and 2) in the 310-helix (Fig. 1b), which has an integer number of amino acids per turn (≅3.0), a triplet of residues selected carefully will produce the expected amphiphilicity.
The amphiphilic helical structure of most antibacterial peptides is a prerequisite for their propensity to form channels across the double-layered biological membranes (12). In aqueous solution, positions a and d of the α-helical heptad repeat (a, b, c, d, e, f, g) (13) require hydrophobic residues for the onset of the widespread antiparallel dimer (or multimer) superstructure (α-helix coiled coil) (14-16) (Fig. 2). The hydrophilic positions e and g, immediately on the back, reinforce the dimer stability via ionic interactions. Deep knowledge of the factors operative in helix dimer formation may help our understanding greatly of protein-protein interactions in chemical biology.
The preferred conformation of peptides based heavily on Ca-ethylated α-amino acids (3) is different. In the resulting 2.05-helical conformation, intramolecular H-bonding takes place between the N-H and C=O groups of the same amino acid building block (17, 18). To achieve the pentagonal disposition, the H-bonding parameters are distorted remarkably and the sp3 N-Ca-C' bond angle is compressed severely (by ≅6.5°). An example of this fully extended (all-traws) conformation, based on Ca,a-diethylglycine (3, R=CH2-CH3) (17), is illustrated in Fig. 3. This twofold structure is extremely rare in globular proteins and is known only for a Gly-rich sequence (19).
Only few Ca-alkylated a-amino acids occur naturally (in peptide antibiotics) (20, 21), which include Aib (α-aminoisobutyric acid or Ca,a-dimethylglycine) (1, R=CH3), Iva (isovaline or Ca-ethyl, Ca-methylglycine) (1, R=CH2-CH3), and (αEt)Nva (Ca-ethylnorvaline) (3, R=(CH2)2-CH3).
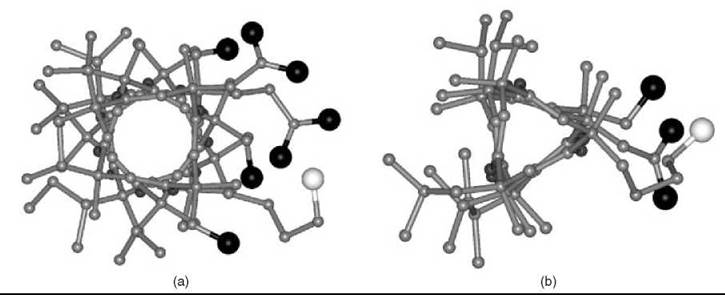
Figure 1. Right-handed (a) α-helical and (b) 310-helical peptide models from L-configurated, Ca-methylated a-amino acids viewed along the helix axis, highlighting their amphiphilic properties. In these models, the largest black (oxygen) and white (nitrogen) atoms on the right side are those of the hydrophilic side chains of Ser, Asp, Glu, and Lys.

Figure 2. Antiparallel dimer formation generated by an amphiphilic α-helix.
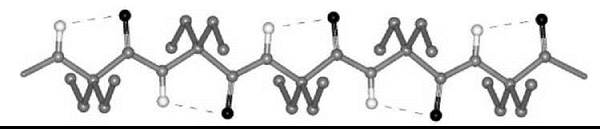
Figure 3. The X-ray diffraction structure of the 2.05-helical homo-pentapeptide from Ca,a-diethylglycine with five, consecutive C=O∙∙∙H-N intramolecular H-bonds (17).
Peptides Based on Ca,β-Didehydro α-Amino Acids
Ca,β-Didehydro α-amino acids (∆AAs) are found in a variety of naturally occurring microbial and fungal metabolites, in a limited number of globular proteins, and in polycyclic peptide antibiotics (22). Bioactive peptide molecules that contain these residues are less prone to enzymatic degradation.
The double bond between the sp2 Ca and Cβ atoms of ∆AAs induces higher lipophilicity and geometric alterations in the bond distances and angles and restricts the conformational flexibilitites of the peptide backbone and the side chain as compared with those of protein residues. Most 3-D structural studies in this subclass of peptidomimetics have exploited ∆Phe mainly because of its convenient chemical synthesis and interesting conformational properties. ∆Phe exists in two diastereomeric forms: the Z-configurational isomer (∆ZPhe, 4), where the N-H group is in the cis disposition with respect to the benzyl moiety, and the E-isomer (∆EPhe, 5), where it is in the trans disposition. Almost all conformational studies described so far have been performed on the Z-isomer (22-24) essentially because a large part of the synthetic routes results in this stereoisomer.
From the investigations on ∆Phe-based peptides, it is clear that the 3-D disposition largely preferred by this residue is the helical conformation (23, 24). Therefore, most corner positions of the various types of β-turns and all positions of 310-/α-helices are accessible easily to this residue. Fig. (4) shows the right-handed 310-helix generated by a double repetition of the model triplet -(∆ZPhe-∆ZPhe-L-Ala)-. Aromatic amino acid residues, which include ∆ZPhe (25), are known to play a significant role in the stability of the helixcoiled coil motifs. The importance of weak interactions that involve aromatic moieties in molecular recognition and in the de novo design of miniproteins has been highlighted frequently.
Another useful property of ∆Phe is its strong absorption in the near-UV region (≅280 nm). This characteristic has been exploited in several studies to probe peptide conformational transitions and helix screw sense changes of chiral switches (26-28).
Interestingly, a series of homo-peptides derived from the Ca,β-didehydro α-amino acid with the shortest side chain (Ca,β-didehydroalanine or ∆Ala, 6) was shown unambiguously to overwhelmingly adopt a multiple, consecutive, fully extended conformation (29). These peptide molecules are essentially flat, including the amino acid side chains, and form completely planar sheets (Fig. 5). This uncommon peptide 3-D structure is stabilized by two types of intramolecular H-bonds, Ni-H∙∙∙Oi=Ci (typical of the 2.05-helix) and Cβi+1-H∙∙∙Oi=Ci (characteristic of ∆Ala peptides). The molecules are isolated and pack in layers without any significant contribution from intermolecular N-H∙∙∙O=C H-bonds. For this exceptionally flat (”sole-like”) peptide structure, it is reasonable to foresee a bright future as a potentially active bridge in charge transfer systems.
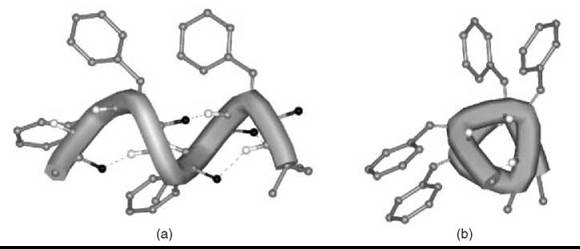
Figure 4. Right-handed 310-helical conformation formed by a double repetition of the -(∆ZPhe-∆ZPhe-L-Ala)- model peptide: views orthogonal to (a) and down (b) the helix axis.
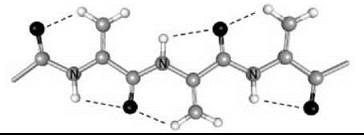
Figure 5. The variant of the 2.05 helical conformation adopted by the homo-tripeptide from Ca,β-didehydroalanine (29).
Peptides Based on N-Substituted α-Amino Acids (Peptoids)
In 1992, Simon et al. (30) introduced a new concept in the search for potentially bioactive peptide molecules: The side chain on the α-amino acid a-carbon is shifted by one atom along the backbone to the next nitrogen to generate a peptoid (7) (i.e., an N-substituted oligoglycine). Peptoids have been shown to be protease resistant (only tertiary amide groups are present in the main chain), oral bioavailable, and easy to synthesize in oligomeric forms with complex sequences (31). Peptoids have been also exploited for the construction of many biologically active compounds in many different fields of pharmaceutical research.
Because of this principal structural divergence from coded peptides, peptoids lack amide protons. This property precludes the formation of the intramolecular H-bonds that contribute largely to the stabilization of the most common helical structures of α-peptides. Unlike that of α-peptides, the peptoid backbone is inherently achiral (as it is based on Gly residues). However, it has been reported that sufficient bias to form stable helices of a specific screw sense can be provided by side chains with an α-chiral carbon atom (that is linked directly to nitrogen) (32, 33). The extraordinary resistance of these helices to loss of their ordered secondary structure is another intriguing property of peptoid molecules.
The only published X-ray diffraction structure for any peptoid oligomer, the Nrch (8) homo-pentamer, shows clearly that the molecule is folded in a left-handed helical conformation with all tertiary amide bonds in the cis conformation (33). The periodicity of this helix is approximately three residues per turn, with a pitch (axial translation per turn) of about 6.7 A. The C=O groups are aligned with the helix axis. The handedness of the peptoid helix is controlled by the chirality of the N-side chain enantiomer. The values of the φ, ψ backbone torsion angles for each residue indicate a semi -extended conformation [i.e., close to the classical type-I poly(Pro)n helix] (Fig. 6).
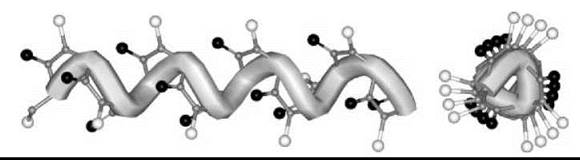
Figure 6. Molecular representation of the left-handed peptoid helix with the φ, ψ, ω backbone torsion angles typical of type-I poly(L-Pro)n.
Peptoid helices are detected in structure-supporting solvents even in relatively short oligomers. Because intramolecular C=O∙∙∙H-N H-bond formation cannot be the driving force for peptoid secondary structure, the steric influence of the bulky and chiral side chain is likely to provide the required constraint. Interactions between side-chain groups, and between side chains and the carbonyls of the main-chain amides, may add stability to the ordered secondary structure. However, for very short oligomers (34) or peptoids based on N-substituted a-amino acids with a small side chain (35), such as Nala (also termed sarcosine, Sar, 9), complex mixtures of conformers associated with either cis or trans tertiary amide groups have been detected. In addition to the classic CD technique, the contribution of other spectroscopies, such as pulsed ESR (36), may be of value for the 3-D structural validation of peptoid molecules.
Peptoid nanostructures will be useful for investigating principles of macromolecule self-assembly and for fabricating novel functional architectures in biotic systems.
Peptides Based on β-Amino Acids
ω-Amino acids (that have more than one carbon atom intervening between the amino and carboxyl functions) possess a larger degree of conformational variability than a-amino acids. Initial conformational investigations of oligomers of to-amino acids were driven by the extensive interest in the various forms of nylons. More recently, a large body of 3-D structural investigations on β-amino acid oligomers have been discovered, but the conformational preferences of peptides based on γ- and δ-amino acids and of “hybrid” peptides (e.g., those formed by α- and β- or α- and γ-amino acids) are also the subject of numerous spectroscopic and X-ray diffraction analyses (37-39). In this section, we summarize the most relevant conformational results extracted from the studies of peptides rich in the β-amino acids listed below.
In β-Ala (10), the simplest amino acid of this class, the central, characteristic C2-C3 bond is unconstrained and can adopt values for the related θ (also called |x in some publications) torsion angle of approximately ±60° (gauche) and 180° (trans). Detailed analyses of the rotational flexibility about this bond in linear β-Ala peptides revealed that θ is mostly trans oriented (40, 41). Also, these studies provided almost exclusive evidence for the formation of extended peptide chains, which usually self-aggregate into sheet-like structures.
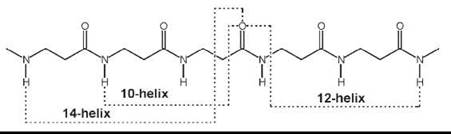
More recently, introduction of (one or multiple) substituents and chiralities on the C2 and C3 atoms (11-13, 15, 16), and incorporation of C2↔C3 (14), N↔C2 (17), and N↔C3 (18) cyclized motifs seemed to facilitate folding of P-peptides into a new type of helices (42-47). A variety of these ordered structures has been authenticated. If the nomenclature is used that terms peptide helices according to the number of atoms involved in the pseudocyclic structure generated by the intramolecular C=O∙∙∙H-N H-bond, those found most frequently are the 14-, 12-, and 12/10-helices. Interestingly, although the direction of the H-bond for the 12-helix is the same as that typical of α-peptides (i.e., from a C-terminal N-H group to an N-terminal C=O group; in this specific case i ← i + 3), the direction is reversed (i → i + 1 and i → i + 2, respectively) for the 10- and 14-helices. β-Amino acids in the extended conformation may also produce parallel or antiparallel pleated-sheet structures.
The 14-helix emerged as the best documented structure among the β-peptide folded structures. The φ, ψ torsion angles are extended (±130 — 140°). The 0 value is constrained (e.g., by the cyclohexyl ring of the cyclic β2,3AA) (14) (n = 2), close to ![]() 60°. Both the pitch and the diameter of this ternary helix are ≅5.0 A.
60°. Both the pitch and the diameter of this ternary helix are ≅5.0 A.
When the substituents on the C2 and C3 atoms of the β-amino acid are part of a five-membered ring (as in the trans cyclic β2,3 AA, 14, with n = 1), the accessible θ torsion angle is restricted to about ±95°. The resulting 12-helix has a set of φ, ψ torsion angles near ![]() 90°. The helix repeats approximately every 2.5 residues, with a pitch of ≅5.5 A, and a diameter slightly less than 4.0 A.
90°. The helix repeats approximately every 2.5 residues, with a pitch of ≅5.5 A, and a diameter slightly less than 4.0 A.
The 12/10-helix was found in alternating β2-(11) and β3-(12) monosubstituted residues. The dipeptide unit that contains a -CONH-bond with no adjacent R2/R3 substituents generates the 12-membered ring, whereas that with the two adjacent substituents is part of the 10-membered ring. This ordered structure consists of consecutive narrower 10-membered and wider 12-membered H-bonded rings. Consequently, the two successive H-bonded rings exhibit opposite directionality. In this conformationally mixed helix, the peptide planes of the 10-atom rings are approximately perpendicular to the helix axis, whereas the peptide planes of the 12-atom rings are aligned nearly with the helix axis. As in the 14-helix, the 0 value is close to ±60°. The number of amino acids per turn is 2.7. The overall helix macrodipole is much smaller compared with those of the other helical structures. The C=O (and the N-H) bonds show an alternating up/down direction with respect to the helix axis.
Two (parallel and perpendicular to the helix axis) views for each of the three β-peptide helices discussed in this section are presented in Fig. 7. Recent applications of β-peptide helices to relevant biological issues (antibacterial activity, transport across cell membranes) (48-51) will certainly stimulate additional work in this promising area.

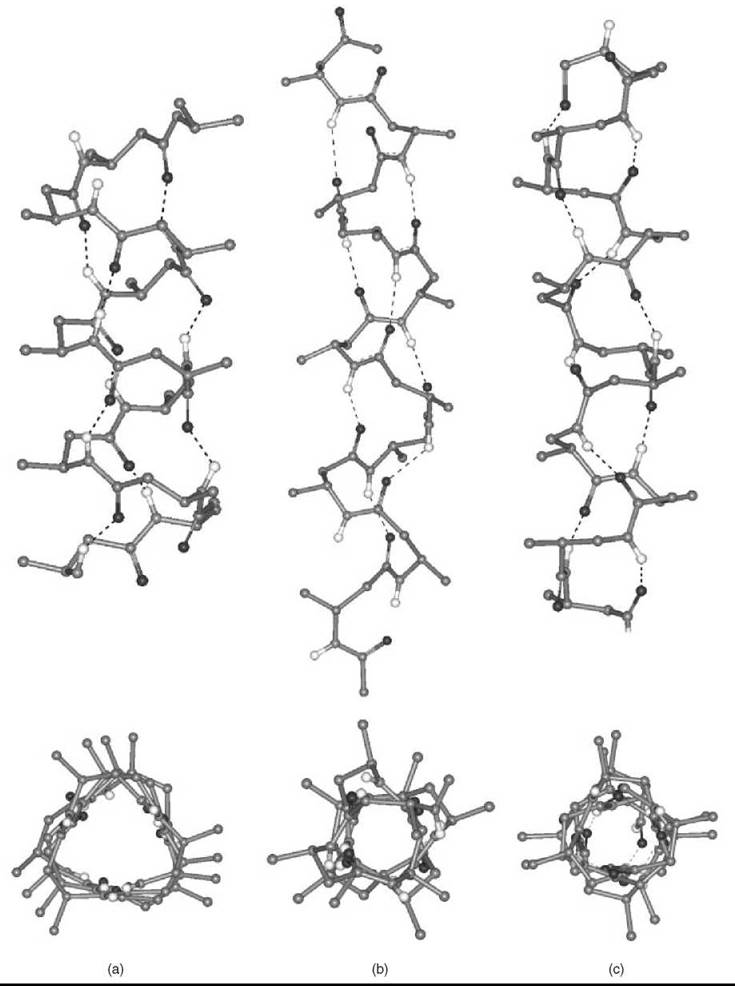
Figure 7. Two orthogonal representations for each of the β-peptide 14-, 12-, and 12/10-helices (a, b, and c, respectively) formed by a β2 AA or/and a β3 AA, where R2/R3 are methyl goups. The H-atoms, except the -CONH- H-atoms, are omitted for clarity.
Lactam-Based Peptides
Several review articles and books have dealt extensively with the synthetic problems encountered and have discussed in detail the biological data of lactam-based peptides. However, insufficient attention has been paid to the conformational implications induced in the resulting constrained analogs (52-56). Here, we will mention briefly only those lactam-based conformational restrictions in peptides that are achieved by short-range cyclizations that involve the backbone N and Ca atoms (from residue i to residue i or i + 1). Therefore, conformational restrictions that involve the C' (carbonyl) atom and those obtained by medium- and long-range cyclizations, although of great theoretic and practical interest, have not been considered. In addition, in all peptides discussed in this section, the repeating N-Ca-C' sequence is maintained.
As a part of a program that evaluates N-acylated γ-lactams as conformationally constrained building blocks of pseudopeptide foldamers, homo-oligomers of L-pyroglutamic acid (L-pGlu) (19) to the tetramer level were synthesized (57). The preferred conformation of this pseudopeptide series with Ni ↔ Cia ring restriction in structure-supporting solvents was assessed by various spectroscopic techniques. In addition, the crystal-state structure of the Na-protected dimer was established by X-ray diffraction. A high-level DFT computational modeling was also performed based on the crystallographic parameters. In this analysis, it was demonstrated that an αC-H∙∙∙O=C intramolecular H-bond is responsible for the stabilization of the s-trans -L-pGlu-L-pGlu- conformation by 1.4 kcal mol-1. This effect can be detected easily by 1H NMR because of the anomalous chemical shifts of the αCH protons present in all oligomers. In summary, a new polyimide-based, foldameric structure (Fig. 8) was developed that, if functionalized appropriately, holds promise as a rigid scaffold for novel functions and applications.

Figure 8. Computer model of the ternary helical structure of the pseudopeptide -(L-pGlu)4-segment generated by the repeating ψ1, ω1, and ψ2 backbone torsion angles obtained from the X-ray diffraction structure of Boc-(L-pGlu)2-OH (Boc, tert-butyloxycarbonyl) (57).
The lactam restriction of peptide conformation (20) (Cia ↔ Ni+1) was proposed first by Freidinger et al. (58) with the aim at forcing the peptide bond (ωi) to be trans and placing a severe constraint on the ψi torsion angle. Conformational energy calculations for (S)-3-amino-2-pyrrolidone (γ-lactam) and (S)-3-amino-2-piperidone (δ-lactam) derivatives indicate that the most stable conformations have ψ1 values restricted to the range -125 ± 10°. As a consequence, these chiral mono-lactams can be accommodated readily at the left corner of a type II’ β-turn (φ = 60°, ψ = -120°) (10, 11), a position occupied typically by a-amino acids of R(D) chirality, but obviously not at the same position of a type-II β-turn (φ = -60°, ψ = 120°). X-ray diffraction, 1H NMR, CD, and computer graphics analyses on a variety of analogs of bioactive peptides are in excellent agreement with the theoretic findings, in particular that underlie the strict requirement of reversing the chirality of the residue at the left-corner of the β-turn to keep the same type (II or II’) of chain folding when a 5(or 6)-membered ring lactam replacement is involved (59). Despite the gross conformational similarities between the 5- and 6-membered ring lactams, distinct biological properties usually are noted for peptides that contain such rings. More subtle differences in conformation and/or in bulkiness between the two lactams might be responsible for the observed diverging biological activities.
The effect of the incorporation of a succinimide (21) moiety into a peptide chain (another type of Cia ↔ Ni+1 ring restriction) has been examined carefully (60, 61). The five-membered succinimide annular system is an intermediate in the repair of selective degradations of side-chain deamidated proteins. It has been shown both theoretically and experimentally that L-Asu-Xxx- (Asu, α-aminosuccinyl) dipeptide sequences strongly favor the type-II’ β-turn, a behavior similar to that of the γ-lactam modification discussed above.
The conformational preference of the terminally blocked homo-trimer from (2S, 4R)-4-amino-5-oxopyrrolidine-2-carboxylic acid (22a), characterized by Cia ↔ Cai+1 ring restriction and an unique, alternating cis-trans secondary amide sequence was analyzed by X-ray diffraction (62). Using computer modeling, it was also shown that the rigid 3-D structure of the trimer can be exploited as a template to construct novel linear oligopeptide foldamers and large-ring cyclic correlates with self-recognizing properties (for the latter, see Fig. 9).

Figure 9. Computer models of: (a) the cyclic correlate based on 12 monomeric units of (2S,4R)-4-amino-5-oxopyrrolidine-2-carboxylic acid. The idealized backbone φ, ψ torsion angles are 120°, 120°, respectively. (b) The nanotubular structure resulting from vertical self-assembling of the cyclic correlate through intermolecular N-H∙∙∙O = C H-bonds that involve the trans amides (62). The black and white atoms are oxygens and hydrogens, respectively.
The most stable conformation of peptides that contain 3-amino-2-piperidone-6-carboxylic acid (22b), also an amino acid with a Cia ↔ Cai+1 ring restriction, has been determined by 1H NMR (63). A chair conformation is a feature that characterizes the piperidone ring, Also, an intramolecularly H-bonded β-turn form is the most plausible explanation for the experimental finding on these peptides.
The synthesis of a novel macrocyclic (10-membered ring) dilactam (23, n = 2), an additional template with a Cia ↔ Cai+1 ring restriction that may promote formation of type-II β-turns, has been reported (64). By analysis of the 1H NMR spectrum, it was established that a chair-like conformation is preferred for this ring structure and that the two amide groups are trans with roughly parallel planes. It seems, however, that the presence of the two macrocyclic secondary amide functions perpendicular to the plane of the turn would facilitate intermolecular association that leads to poor solubility and gel formation. A similar (11-membered) ring structure has been synthesized to generate a relatively rigid model of the bioactive peptide thymopentin. Peptides that contain two consecutive macrocyclic dilactams, separated by a Ca-C'-N-Ca region that retains flexibility about the ψi+1 and ψi+2 torsion angles, have been prepared (65). This swivel-like effect can allow the peptides to adopt low-energy folded conformations. From potential energy calculations, it was established that the best candidates for β-turn models are the 12-membered ring structures that have the configurational sequence L,L,D,D (in these compounds all amide groups are nearly trans planar). The 10- and 11-membered dilactams were shown to be higher in energy. The results of a CD-1H NMR study are consistent with the presence of a β-turn in the —L-Lys-L-Glu-D-Lys-D-Glu-Ms-dilactam-bridged tetrapeptide.
In search for enkephalinamides with improved analgesic activity, conformationally restricted analogs, where the Tyr, Gly, Phe, and Leu/Met residues are cyclized to form a piperazin-2-one (24) (an Ni ↔ Ni+1 ring restriction), have been prepared. Analogs with either D-Tyr1 or L-Leu5(L-Met5) are active. The absolute configuration of the piperazin-2-one derived from the intermediate dipeptide amide H-L-Phe-L-Leu-NH2 has been assessed by X-ray diffraction. The enantioface-differentiating abilities of cyclic peptides that contain N,N’-ethylene-bridged dipeptides have been examined. The X-ray diffraction structure of an L-Ala-L-Ala piperazin-2-one derivative has been described (66). The ψi torsion angle is fixed at about —35°, whereas фi and ωi, in part external to the ring structure, are trans.

Summary
In this introductory review we have presented the most relevant 3-D structural characteristics of a selected set of peptidomimetics that, hopefully, will be of help to the nonspecialized scientists for potential use in the currently exploding field of chemical biology. More specifically, we have discussed examples of peptide surrogates with main-chain and side-chain modifications either at the α-nitrogen or at the α-carbon, or at both α- and β-carbons, or with different types of cyclizations (N↔N, N↔Ca, and Ca↔Ca). For journal issues or books devoted to these and other types of peptidomimetics, the reader is referred to References 67-73.
References
1. Spatola A. Peptide backbone modifications. A structure-activity analysis of peptides containing amide bond surrogates: conformational constraints and related amide bond replacements. In: Chemistry and Biochemistry of Amino Acids, Peptides, and Proteins: A Survey of Recent Developments, Vol. 7. Weinstein B, ed. 1983. Marcel Dekker Inc., New York. pp. 267-357.
2. Houben-Weyl. Methods of Organic Chemistry, vol. E22c. Goodman M, Felix A, Moroder L, Toniolo C, eds. 2003. Georg Thieme Verlag, Stuttgart, Germany.
3. Giannis A, Kolter T. Peptidomimetics for receptor ligands. Discovery, development, and medical perspectives. Angew. Chem. Int. Ed. Engl. 1993; 32:1244-1267.
4. Gante J. Peptidomimetics. Tailored enzyme inhibitors. Angew. Chem. Int. Ed. Engl. 1994; 33:1699-1720.
5. Marshall GR. Studies of the biologically active conformations of angiotensin, Vol 5. In: Intra-Science Chemistry Reports. Kharasch N, ed. 1971. Gordon and Breach Science Publishers, New York. pp. 305-316.
6. Karle IL, Balaram P. Structural characteristics of α-helical peptide molecules containing Aib residues. Biochemistry 1990; 29:6747- 6756.
7. Toniolo C, Benedetti E. The polypeptide 310-helix. Trends Biochem. Sci. 1991; 16:350-353.
8. Bolin KA, Millhauser GL. α and 310: the split personality of polypeptide helices. Acc. Chem. Res. 1999; 32:1027-1033.
9. Toniolo C, Crisma M, Formaggio F, Peggion C. Control of peptide conformation by the Thorpe-Ingold effect (Ca-tetrasubstitution): Biopolymers (Pept. Sci.) 2001; 60:396-419.
10. Venkatachalam CM. Stereochemical criteria for polypeptides and proteins. V. Conformation of a system of three linked peptide units. Biopolymers 1968; 6:1425-1436.
11. Rose GD, Gierasch LM, Smith JA. Turns in peptides and proteins. Adv. Protein Chem. 1985; 37:1-109.
12. Sansom MSP. The biophysics of peptide models of ion channel. Progr. Biophys. Mol. Biol. 1991; 55:139-235.
13. Kwok SC, Hodges RS. Effect of chain length on coiled-coil stability: decreasing stability with increasing chain length. Biopolymers (Pept. Sci.) 2004; 76:378-390.
14. DeGrado WF, Summa CM, Pavone V, Nastri F, Lombardi A. De novo design and structural characterization of proteins and metalloproteins. Annu. Rev. Biochem. 1999; 68:779-819.
15. Liu J, Zheng Q, Cheng CS, Kallenbach NR, Lu M. A seven-helix coiled coil. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:15457-15462.
16. Hadley EB, Gellman SH. An antiparallel α-helical coiled-coil model system for rapid assessment of side-chain recognition at the hydrophobic surface. J. Am. Chem. Soc. 2006; 128:16444-16445.
17. Benedetti E, Barone V, Bavoso A, Di Blasio B, Lelj F, Pavone V, Pedone C, Bonora GM, Toniolo C, Leplawy MT, Kaczmarek K, Redlinski AS. Structural versatility of peptides from Caa,-dialkylated glycines. I. A conformational energy computation and X-ray diffraction study of homo-peptides from Ca,a- diethylglycine. Biopolymers 1988; 27:357-371.
18. Imawaka N, Tanaka M, Suemune H. The first fully planar C5-conformation of homo-oligopeptides prepared from a chiral α-ethylated α,α-disubstituted amino acid: (S)-butylethylglycine [= (2S)-2-amino-ethylhexanoic acid]. Helv. Chim. Acta 2000; 83:2823-2835.
19. Aberg A, Yaremchuk A, Tukalo M, Rasmussen B, Cusack S. Crystal structure analysis of the activation of histidine by Thermus thermophilus histidyl-tRNA synthetase. Biochemistry 1997; 36:3084-3094.
20. Toniolo C, Crisma M, Formaggio F, Peggion C, Epand RF, Epand RM. Lipopeptaibols, a novel family of membrane active, antimicrobial peptides. Cell. Mol. Life Sci. 2001; 58:1179-1188.
21. Rainaldi M, Moretto A, Peggion C, Formaggio F, Mammi S, Peggion E, Galvez JA, Diaz-de-Villegas MD, Cativiela C, Toniolo C. Lipopeptaibol metabolites of Tolypocladium geodes: total synthesis, preferred conformation, and membrane activity. Chem. Eur. J. 2003; 9:3567-3576.
22. Stammer CH. Dehydroamino acids and peptides, Vol. 6. In: Chemistry and Biochemistry of Amino Acids, Peptides and Proteins. Weinstein, Boris, ed. 1982. Marcel Dekker, New York. pp. 33-74.
23. Singh TP, Kaur P. Conformation and design of peptides with α,β-dehydroamino acid residues. Progr. Biophys. Mol. Biol. 1996; 66:141-165.
24. Jain R, Chauhan VS. Conformational characteristics of peptides containing α,β-dehydroamino acid residues. Biopolymers (Pept. Sci.) 1996; 40:105-119.
25. Ramagopal UA, Ramakumar S, Mathur P, Joshi R, Chauhan VS. Dehydrophenylalanine zippers: strong helix-helix clamping through a network of weak interactions. Protein Eng. 2002; 15:331-335.
26. Pieroni O, Fissi A, Pratesi C, Temussi PA, Ciardelli F. Reversible screw sense inversion of the 310-helix in a dehydropeptide. J. Am. Chem. Soc. 1991; 113:6338-6340.
27. Pieroni O, Fissi A, Jain RM, Chauhan VS. Solution structure of dehydropeptides: a CD investigation. Biopolymers 1996; 38:97-108.
28. Komori H, Inai Y. Control of peptide helix sense by temperature tuning of non-covalent chiral domino effect. J. Org. Chem. 2007; 72:4012-4022.
29. Crisma M, Formaggio F, Toniolo C, Yoshikawa T, Wakamiya T. Flat peptides. J. Am. Chem. Soc. 1999; 121:3272-3278.
30. Simon RJ, Kania RS, Zuckermann RN, Huebner VD, Jewell DA, Banville S, Ng S, Wang L, Rosenberg S, Marlowe CK, Spellmeyer DC, Tan R Frankel AD, Santi DV, Cohen FE, Bartlett PA. Peptoids: a modular approach to drug discovery. Proc. Natl. Acad. Sci. U.S.A. 1992; 89:9367-9371.
31. Richter LS, Spellmeyer DC, Martin EJ, Figliozzi GM, Zuckermann RN. Automated synthesis of nonnatural oligomer libraries: the peptoid concept. Combinatorial Peptide and Nonpeptide Libraries.Jung G, ed. 1996. VCH, Weinheim, Germany. pp. 387-404.
32. Sanborn TJ, Wu CW, Zuckermann RN, Barron AE. Extreme stabilities of helices formed by water-soluble poly-N-substituted glycines (polypeptoids) with α-chiral side chains. Biopolymers 2002; 63:12-20.
33. Wu CW, Kirshenbaum K, Sanborn TJ, Patch JA, Huang K, Dill KA, Zuckermann RN, Barron AE. Structural and spectroscopic studies of peptoid oligomers with α-chiral aliphatic side chains. J. Am. Chem. Soc. 2003; 125:13525-13530.
34. Rainaldi M, Moretto V, Crisma M, Peggion E, Mammi S, Toniolo C, Cavicchioni G. Peptoid residues and p-turn formation. J. Pept. Sci. 2002 ;8:241-252.
35. Toniolo C, Bonora GM, Schilling FC, Bovey FA. Proton magnetic resonance study of linear sarcosine oligomers. Macromolecules 1980; 13:1381-1385.
36. Fafarman AT, Borbat PT, Freed JH, Kirshenbaum K. Characterizing the structure and dynamics of folded oligomers. Pulsed ESR studies of peptoid helices. Chem. Commun. 2007; 377-379.
37. Roy RS, Balaram P. Conformational properties of hybrid peptides containing α- and ω-amino acids. J. Pept. Res. 2004; 63:279-289.
38. Baldauf C, Gunther R, Hofmann HJ. Helix formation in α,γ- and β,γ-hybrid peptides: theoretical insights into mimicry of α- and β-peptides. J. Org. Chem. 2006; 71:1200-1208.
39. Price JL, Horne WS, Gellman SH. Discrete heterogeneous quaternary structure formed by α/β-peptide foldamers and α-peptides. J. Am. Chem. Soc. 2007; 129:6376-6377.
40. Banerjee A, Balaram P. Stereochemistry of peptides and polypeptides containing w-amino acids. Curr. Sci. 1997; 73:1067-1077.
41. Kishore R. p-Ala containing peptides: potentials in design and construction of bioactive peptide and protein secondary structure mimics. Curr. Protein Pept. Sci. 2004; 5:435-455.
42. DeGrado WF, Schneider JP, Hamuro Y. The twists and turns of β-peptides. J. Pept. Res. 1999; 54:206-217.
43. Cheng RP, Gellman SH, DeGrado WF. β-Peptides: from structure to function. Chem. Rev. 2001; 101:3219-3232.
44. Martinek TA, Fulop F. Side-chain control of β-peptide secondary structures. Design principles. Eur. J. Biochem. 2003; 270:3657- 3666.
45. Seebach D, Beck AK, Bierbaum DJ. The world of β- and γ-peptides comprised of homologated proteinogenic amino acids and other components. Chem. Biodiv. 2004; 1:1111-1239.
46. Seebach D, Kimmerlin T, Sebesta R, Campo MA, Beck AK. How we drifted into peptide chemistry and where we have arrived at. Tetrahedron 2004; 60:7455-7506.
47. Seebach D, Hook DF, Glattli A. Helices and other secondary structures of β- and γ-peptides. Biopolymers (Pept. Sci.) 2006; 84:23-27.
48. Hamuro Y, Schneider JP, DeGrado WF. De novo design of antibacterial β-peptides. J. Am. Chem. Soc. 1999; 121:12200-12201.
49. Porter EA, Weisblum B, Gellman SH. Mimicry of host-defense peptides by unnatural oligomers: antimicrobial β-peptides. J. Am. Chem. Soc. 2002; 124:7324-7330.
50. Umezawa N, Gelman MA, Haigis MC, Raines RT, Gellman SH. Translocation of a β-peptide across cell membranes. J. Am. Chem. Soc. 2002; 124:368-369.
51. Rueping M, Mahajan Y, Sauer M, Seebach D. Cellular uptake studies with β-peptides. ChemBioChem. 2002; 3:257-259.
52. Toniolo C. Conformationally restricted peptides through shortrange cyclizations. Int. J. Pept. Protein Res. 1990; 35:287-300.
53. Ball JB, Alewood PF. Conformational constraints: nonpeptide β-turn mimics. J. Mol. Recogn. 1990; 3:55-64.
54. Holzemann G. Peptide conformation mimetics (part 1). Kontakte (Darmstadt) 1991; 3-12.
55. Liskamp RMJ. Conformationally restricted amino acid and dipeptides, (non)peptidomimetics and secondary structure mimetics. Rec. Trav. Chim. Pays-Bas 1994; 113:1-19.
56. Gillespie P, Cicariello J, Olson GL. Conformational analysis of dipeptide mimetics. Biopolymers 1997; 43:191-217.
57. Bernardi F, Garavelli M, Scatizzi M, Tomasini C, Trigari V, Crisma M, Formaggio F, Peggion C, Toniolo C. Pseudopeptide foldamers: the homo-oligomers of pyroglutamic acid. Chem. Eur. J. 2002; 8:2516-2525.
58. Freidinger RM, Veber DF, Hirschmann R, Paege LM. Lactam restriction of peptide conformation in cyclic hexapeptides which alter rumen fermentation. Int. J. Pept. Protein Res. 1980; 16:464-470.
59. Valle G, Crisma M, Toniolo C, Yu KL, Johnson RL. Crystal-state structural analysis of two γ-lactam restricted analogs of Pro-Leu-Gly-NH2. Int. J. Pept. Protein Res. 1989; 33:181-190.
60. Capasso S, Mattia CA, Mazzarella L, Zagari A. Conformational properties of aminosuccinyl peptides. Crystal structure and conformational analysis of tert-butyloxycarbonyl-L-aminosuccinyl- glycine methyl ester. Int. J. Pept. Protein Res. 1984; 23:248-255.
61. Capasso S, Mazzarella L, Sica F, Zagari A. Chiroptical properties of aminosuccinyl peptides. Int. J. Pept. Protein Res. 1989; 33:124-132.
62. Crisma M, Moretto A, Toniolo C, Kaczmarek K, Zabrocki J. First rigid peptide foldamers with an alternating cis-trans amide sequence. An oligomeric building block for the construction of new helices, large-ring cyclic correlates, and nanotubes. Macromolecules 2001; 34:5048-5052.
63. Kemp DS, McNamara P. Amino acid derivatives that stabilize secondary structures of polypeptides. II. The most stable conformation of peptides containing 3-amino-2-piperidone-6-carboxylic acid (Acp). Tetrahedron Lett. 1982; 23:3761-3764.
64. Kemp DS, Stites WE. A convenient preparation of derivatives of 3(S)-amino-10(R)-carboxy-1,6-diaza-cyclodeca-2,7-dione, the dilactam of L-α,γ-diaminobutyric acid and D-glutamic acid: a β-turn template. Tetrahedron Lett. 1988; 29:5057-5060.
65. Rone R, Manesis N, Hassan M, Goodman M,. Hagler AT, Kitson DH, Roberts VA. Conformational analysis of constrained dilactam-bridged tetrapeptides. Tetrahedron 1988; 44:895-924.
66. Yamashita T, Kojima Y, Hirotsu K, Ohsuka A. Macrocyclic peptides. Synthesis and structure of a novel dipeptide, (2S, 3’S)-2-(2’-oxo-3’-methylpiperazin-1’-oxyl)-propanoic acid, and its use as the unit of cyclic peptides. Int. J. Pept. Protein Res. 1989; 33:110-114.
67. Rizo J, Gierasch LM. Constrained peptides: model of bioactive peptides and protein substructures. Annu. Rev. Biochem. 1992; 61:387-418.
68. Marraud M, Aubry A. Crystal structures of peptides and modified peptides. Biopolymers (Pept. Sci.) 1996; 40:45-83.
69. Hanessian S, McNaughton-Smith G, Lombart HG, Lubell WD. Design and synthesis of conformationally constrained amino acids as versatile scaffolds and peptide mimetics. Tetrahedron (report No. 426) 1997; 53:12789-12854.
70. Peptides and Peptidomimetics that Adopt Folded Structures. Bioorg. Med. Chem. 1999;7, symposia-in-print No. 15.
71. Abell A. Advances in Amino Acid Mimetics and Peptidomimetics, Vol. 2. 1999. JAI Press Inc., Stamford, CT.
72. Kazmierski WM. Peptidomimetics Protocol. Methods in Molecular Medicine. 1999. Humana Press, Totowa, NJ.
73. Hruby VJ. Design in topographical space of peptide and peptidomimetic ligands that can affect behavior. A chemist’s glimpse at the mind-body problem. Acc. Chem. Res. 2001; 34:389-397.
Further Reading
Bayley PD. An Introduction to Peptide Chemistry. 1990. Wiley, Chicester, U.K.
Benoiton NL. Chemistry of Peptide Synthesis. 2005. CRC Press, Boca Raton, FL.
Bodanszky M. Peptide Chemistry: A Practical Textbook. 1988. Springer Verlag, New York.
Gorske BC, Jewell GA, Guerard EJ, Blackwell HE. Expedient synthesis and design strategies for new peptoid construction. Org. Lett. 2005; 7:1521-1524.
Grant G. Synthetic Peptides: A User’s Guide. 2002. Oxford University Press, Oxford.
Gutte B. Peptides: Synthesis, Structures, and Applications. 1995. Academic Press, San Diego, CA.
Howl J. Peptide Synthesis and Applications, Vol. 298. Methods in Molecular Biology. 2005. Humana Press, Totowa, NJ.
Humphrey JM, Chamberlin AR. Chemical synthesis of natural product peptides: coupling methods for the incorporation of noncoded amino acids into peptides. Chem. Rev. 1997; 97:2243-2266.
Jones J. The Chemical Synthesis of Peptides. 1991. Clarendon Press, Oxford.
Noda K, Shimohigashi Y, Izumiya N. α,β-Dehydroamino acids and peptides, Vol. 5. The Peptides: Analysis, Synthesis, Biology. Gross E, Meienhofer J, eds. 1983. Academic Press, New York. pp. 285-339.
Shin SBY, Yoo B, Todaro LJ, Kirshenbaum K. Cyclic peptoids. J. Am. Chem. Soc. 2007; 129:3218-3225.
Zuckermann RN. Efficient method for the preparation of peptoids [oligo(N-substituted glycines)] by submonomer solid-phase synthesis. J. Am. Chem. Soc. 1992; 114:10646-10647.
Zuckermann RN. The chemical synthesis of peptidomimetics. Curr. Opin. Struct. Biol. 1993; 3:580-584.
See Also
Amino Acids, Chemistry of
Natural and Unnatural Amino Acids, Synthesis of
Peptide Synthesis
Peptides, Chemistry of
Synthetic Peptides to Define Structure-Function Relationships