CHEMICAL BIOLOGY
Microarrays in Chemical Biology
Angela N. Koehler, Broad Institute of Harvard, Cambridge, Massachusetts, and MIT, Cambridge, Massachusetts
doi: 10.1002/9780470048672.wecb645
Advances in both genomics and proteomics have provided researchers with access to large collections of biomolecules, including DNA, proteins, and metabolites. High-throughput methods are needed to study the function and regulation of these biomolecules within complex systems. Microarrays have emerged as a common platform to study biomolecular interactions that involve nucleic acids, proteins, and small molecules. DNA microarrays have revolutionized genomic research by allowing researchers to study gene expression, sequence variation, and transcription factor binding sites on a whole-genome scale. Protein microarrays can be used to study interactions with other proteins, DNA, RNA, and small molecules, including lipids, carbohydrates, and drugs. Protein microarrays can also be used as analytical tools to profile complex protein mixtures, such as fractionated cell lysates, in an effort to study antibody specificity, measure changes in protein abundance, or characterize disease states. Small-molecule microarrays are useful tools for ligand discovery, comparing inhibitor specificity across enzyme classes, high-throughput, cell-based phenotypic screens, and as diagnostic tools for pathogen detection. The microarray approach has been extended to transfected cell microarrays, RNAi living cell microarrays, virus microarrays, and tissue microarrays. This article reviews chemical strategies for making microarrays and applications of microarrays in chemical biology.
As the molecular parts list for cells and organisms continues to grow, a key challenge is to identify the function of each component within the context of the cellular system. Increasingly, researchers are choosing to adopt high-throughput, discovery-oriented studies aimed at systems of biomolecules in addition to traditional reductionist studies of the structure or function of a specific biomolecule. Studies of complex biologic systems benefit from large-scale and global analyses as the interacting parts can be analyzed simultaneously, which leads to hypotheses about how the components come together to effect processes such as development, disease, or evolution. Several high-throughput methods have been developed to study the function and regulation of biomolecules, including nucleic acids, proteins, and metabolites. In particular, microarray technology has emerged as a revolutionary platform to analyze all types of biomolecules in a highly parallel fashion.
Biologic Background
Microarrays were introduced in the 1990s for use in genomic studies (1-4). Microscopic features of probe nucleic acids, including oligonucleotides, cDNAs, or PCR products that correspond to predicted or known mRNAs, are deposited on a stable, solid substrate such as glass or silicon. Taking a cue from the Southern blot, the affixed probes are hybridized with cDNA from the sample of interest. Thousands of probes can be accommodated on a single microarray providing an opportunity to interrogate whole genomes (5). DNA microarrays, otherwise known as DNA chips, are valuable tools for gene expression profiling, comparative genomic hybridization (6), genotyping (7), chromatin immunoprecipitation (8), and in vitro studies of protein-DNA interactions (9, 10). DNA microarray technology has also had a significant impact on medicine by facilitating connections between physiologic states and gene expression patterns in studies of cancers, disease progression, and cellular responses to toxins or therapeutics (11). DNA microarray technology is now widely accessible to researchers through commercial microarray products and academic genome centers.
The miniaturized and highly parallel microarray format has also become a common platform in applications that involve other types of biomolecules (Table 1) (1-78). Shortly after the advent of DNA microarrays, small molecules and proteins were captured in the microarray format (12, 13). Microarrays of carbohydrates (14, 15), lipids (16), peptides (17), cells (18), viruses (19), and tissues (20) have also been reported. Why has the microarray format become so attractive for studying diverse types of biomolecules? Assay miniaturization is a key advantage of microarray-based assays. Miniaturization reduces the amount of sample required for the assay and increases the density of samples that may be analyzed simultaneously. Microarrays are also easy to manufacture and store. Most types of microarrays are stable for up to six months under proper storage conditions. Benefiting from advances in automation, replicate arrays can be processed in parallel, which allows researchers an opportunity to evaluate thousands of probes across a variety of sample conditions simultaneously. These advantages make the microarray format attractive to researchers in chemical biology with an interest in studying responses of transcriptomes or proteomes to small molecules. Additionally, microarrays provide an attractive format to screen precious small molecules, such as natural products and compounds coming from combinatorial libraries, against collections of nucleic acids, proteins, or cells. Advances in microarray-related technologies, including new immobilization strategies and detection methods, have lead to novel applications in discovery chemistry (21) and diagnostics (22). These advances will be reviewed in the context of applications related to chemical biology.
Table 1. Summary of microarray types and their applications in chemical biology
|
Microarray type |
Arrayed feature |
Applications |
References |
|
Nucleic acid |
oligos, RNA, DNA |
gene expression profiling, genotyping, mapping protein-DNA interactions, ligand discovery, reaction discovery, RNAi screens |
1-11, 24, 36, 38, 49-51, 76, 77 |
|
Small molecule |
carbohydrates, lipids, natural products, drugs, drug-like molecules |
synthesis platform, ligand discovery, antibody binding screens, enzymatic assays, phenotypic cell-based screens, diagnostics, target deconvolution |
12, 14-16, 21, 22, 25-32, 39, 40, 43, 44, 46, 48, 53, 55-65, 78 |
|
Protein |
antibodies, peptides, protein domains, functional proteins, clarified cell lysates |
enzymatic assays, mapping protein-DNA interactions, mapping protein-protein interactions, mapping post-translational modifications, target deconvolution, antibody, binding screens, protein expression profiling and biomarker discovery |
13, 17, 23, 33-35, 37, 38, 41, 42, 45, 47, 66-75 |
|
Other |
cells, viruses, tissues |
phenotypic screens, RNAi, pathology |
18-20, 22, 52, 54 |
Strategies for Preparing Microarrays and Methods for Detecting Interactions
The first step of any microarray experiment involves design and fabrication of the chips containing probe molecules of interest. Immobilization methods should take both orientation of display and molecular stability into account. Stability is an especially important concern in the area of protein microarrays as many applications require that individual proteins retain their conformation and function (23). Most types of arrays are prepared by immobilizing the biomolecules on chemically treated glass microscope slides using either microcontact spotters or piezoelectric deposition. Microlithography, using either masks or optical methods, is another common method for fabricating microarrays and is used routinely to prepare oligonucleotide arrays (3, 24). Several capture strategies have been developed for microarrays and include both covalent attachment of the probes to surface and noncovalent deposition (Table 2). Both approaches have proven useful in making arrays of nucleic acids, small molecules, and proteins.
Covalent capture strategies
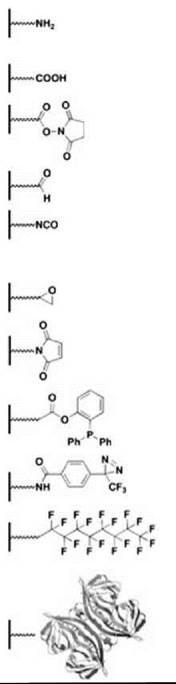
It is common for microarrays to include biomolecules that are attached covalently to a glass or silicon surface. For spotted microarrays, biomolecules are coupled using a variety of attachment chemistries (Table 2). Most advances in attachment chemistry have been driven by the preparation of small-molecule microarrays (SMMs) as small organic molecules tend to have more diverse functional groups than biopolymers. Many attachment chemistries have been reported and reviewed elsewhere (25, 26). Most of these approaches involve mild and selective coupling reactions. The first SMMs made use of a Michael addition reaction that involves molecules containing free thiols printed onto slides coated with vinyl sulfone or maleimide groups (12). Although this strategy proved successful, it was not general as most compound libraries do not contain a high proportion of free thiols. Many more compounds that come from both combinatorial libraries and natural product collections contain amino groups. Although these compounds may be coupled to carboxy-modified glass via amide bond formation, Chang and coworkers (27) chose to print a library of amine-containing molecules on slides coated with N-hydroxysuccinimide (NHS) activated esters. This approach obviates the need for additional coupling catalysts. Readily commercially available epoxide-coated glass slides have been used to capture hydrazide-tagged small molecules and carbohydrates (28, 29). Most surface capture methods take advantage of a reactive functional group that is introduced as part of their synthesis and biases the orientation of the small molecule on the surface. Motivated by the need to increase molecular diversity on SMMs, nonselective approaches to capturing compounds have been adopted. Bradner et al. (30) reported the use of an isocyanate-mediated capture strategy to print nearly 10,000 known bioactive small molecules, natural products, and small molecules originating from several diversity-oriented syntheses. Isocyanates react with a variety of nucleophilic functional groups, thereby increasing the number of small molecules, from natural or synthetic sources, that may be printed on a single surface. Kanoh et al. (31) prepared microarrays of approximately 2000 natural products and drugs by photo-cross-linking compounds on trifluoromethylaryldiazarine-coated surfaces. Using this approach, photogenerated carbenes react with the printed compounds in a manner that is independent of the functional group. Both the isocyanate and the photo-cross-linking strategies present the possibility of printed compounds that occupy multiple modes of orientation within a given spot, which effectively increases the number of binding modes that a given probe protein can sample. Several surface capture methods applied to SMMs have also been used to prepare protein microarrays, including the use of epoxide-coated slides and attachment via Staudinger ligation (32, 33). Additional attachment chemistries have been developed in the context of fabricating protein microarrays and have been reviewed previously (34, 35). Commonly, aldehyde-coated and epoxy-coated slides have been used to capture covalently proteins in a heterogenous fashion by reacting with amino groups (13, 35). As mentioned, retaining the protein function is a key concern when using covalent chemistry to prepare microarrays, and thus, several groups have pursued noncovalent fabrication of functional protein microarrays.
Noncovalent fabrication methods have been used to make microarrays of nucleic acids, small molecules, and proteins (Table 2). Slides coated with aminosilane or poly-L-lysine have been used to capture randomly oligonucleotides (36), proteins (37), and cells (18) via electrostatic interactions or passive adsorption. Similarly, DNA, proteins, and carbohydrates have been arrayed on nitrocellulose-based substrates (38-40). Homogenous orientation may be achieved using affinity tags. For example, large collections of His-tagged proteins have been printed on nickel-coated slides (41). Alternatively, probe biomolecules can be biotinylated and printed on streptavidin-coated surfaces (42). Taking advantage of the highly specific fluorous affinity interaction, Pohl and coworkers (43) noncovalently captured fluorous-tagged carbohydrates on fluoroalkylsilane-coated slides. Winssinger and coworkers (44) prepared SMMs containing a PNA-encoded tetrapeptide acrylate library via sequence-specific hybridization to an oligonucleotide microarray. This elegant approach allows encoding combinatorial libraries and immobilization on-array via self-assembly. Finally, both microwells and microdroplets have been adapted to the microarray format in an effort to carry out experiments in solution (45, 46).
Table 2. Representative capture strategies for preparing microarrays
|
Surface |
Coupling partner |
Linkage type |
Microarray type |
|
|
|
amino |
carboxylic acid, activated ester, epoxy, aldehyde |
covalent or electrostatic, amide, alkylamine, Schiff base |
nucleic acid, small molecule, protein, cell, virus |
|
carboxylic acid |
amine, alcohol |
covalent or electrostatic, amide, ester |
small molecule, protein |
|
|
activated ester |
amine |
covalent, amide |
nucleic acid, small molecule, protein |
|
|
aldehyde |
amine |
covalent, Schiff base |
nucleic acid, small molecule, protein |
|
|
isocyanate |
amine, thiol, hydroxamic acid, alcohol, carboxylic acid |
covalent, urea, carbamothiolate, carbamate, carbamic anhydride |
nucleic acid, small molecule |
|
|
epoxy |
amine alcohol, hydrazide |
covalent, alkylamine, ether |
nucleic acid, small molecule, protein |
|
|
maleimide |
thiol |
covalent, thioether |
nucleic acid, small molecule, protein |
|
|
phosphine |
azide |
covalent, amide |
small molecule, protein |
|
|
diazirine |
nonselective |
covalent, photoaffinity capture |
small molecule |
|
|
fluorous |
C8F17 tag |
noncovalent, fluorophilic interaction |
nucleic acid, small molecule, peptide |
|
|
streptavidin |
biotin tag |
noncovalent, high affinity protein-ligand complex |
nucleic acid, small molecule, protein |
|
Detection methods
Another important planning step involves choice of detection method. Whether using DNA microarrays in whole-genome expression profiling or using small-molecule microarrays to identify new ligands for a protein target of interest, some method of detection is required to locate probe-sample interactions. Typically, probes are labeled with fluorescent, chemiluminescent, radioactive, or affinity tags. Most applications involve a fluorescent readout and the use of a fluorescent microarray slide scanner. A variety of fluorescent reagents, including reactive dyes, fluor-labeled proteins, and fluor-labeled secondary antibodies, are commercially available and considered safer than radioactive labels. Detection strategies may include combinations of different label types. For example, a microarray may be incubated with biotinylated protein followed by incubation with fluorescently labeled streptavidin. Potential disadvantages of label-based detection include overlabeling of protein samples that results in loss of protein conformation or activity. Care should be taken not to label a critical functional group in a small molecule or protein required to bind its target probe. Label-free approaches are gaining momentum in the field of microarrays (47). In particular, surface plasmon resonance (SPR) detection has been used successfully to detect interactions of proteins applied to small-molecule microarrays (48) and will likely prove useful in high-throughput studies that involve protein microarrays.
Applications of Microarrays in Chemical Biology
Microarrays have served as tools for investigations in chemical biology. DNA microarrays are used to study the effects of small molecules on gene expression (49-51). Small-molecule microarrays are used for ligand discovery and enzyme specificity profiling (14-17, 26) as well as for high-throughput, cell-based phenotypic screens (52) and as diagnostic tools (22, 53). Protein microarrays are used for studying interactions with other proteins, nucleic acids, and small molecules, including lipids, carbohydrates, and drugs (23). Protein microarrays are also used as analytical tools to profile complex protein mixtures, such as fractionated cell lysates, in an effort to study antibody specificity, measure changes in protein abundance, or characterize disease states. The microarray approach has been extended to transfected cell microarrays (18), RNAi living cell microarrays (52), virus microarrays (19), and tissue microarrays (20), all of which may be future tools for characterizing the effects of small molecules. A brief sampling of representative applications of microarrays in chemical biology is presented.
Microarrays of nucleic acids
DNA microarrays enable genome-wide measurement of transcriptional responses to small molecule treatment. Typically, drugs and bioactives coming from target-oriented assays or phenotypic cellular assays are profiled in an effort to gain insight into mechanism of action (49). While a great deal of information is gained from these experiments, it is still difficult to draw conclusions about mechanism within isolated data sets. Rather, systematic and comparative analyses using databases of transcriptional profiles have proven valuable in this regard. Lamb et al. (50) described The Connectivity Map as a publicly accessible reference collection of transcriptional profiles in human cultured cells treated with small molecules, including FDA-approved drugs and bioactive tool compounds. By comparing the profile of a novel small molecule to those of known therapeutics, compounds may be annotated as resembling compounds from a known therapeutic class or novel in genomic signature. Additionally, the Connectivity Map project aims to describe genomic signatures of physiological and disease states. In combination with pattern-matching tools, the authors make connections between drugs, genes, and diseases. Specifically, genomic signatures were used to recognize drugs with common mechanisms of action, discover unknown mechanisms, and identify new compounds with therapeutic potential. The authors proposed expanding this resource by profiling all FDA-approved drugs and inhibitory RNAs targeting a wide selection of genes in a panel of diverse cell lines. Individual researchers may perform their own Connectivity Map analyses by using a web-based tool (www.broad.mit.edu/cmap). In an effort to create target identification hypotheses for small molecules positives in phenotypic screens, Butcher et al. (51) used DNA microarrays to monitor effects of gene overexpression on yeast growth in the presence of small molecules. A collection of roughly 3900 Saccharomyces cerevisiae strains harboring different overexpression plasmids was monitored for changes in growth in response to treatment with a small molecule. As a proof of concept, the authors identified genes that, when overexpressed, affect yeast growth in the presence of the natural product drug rapamycin. Target of rapamycin protein (TOR) was successfully identified as candidate rapamycin target and several new genes were implicated in the TOR pathway. The authors used the same microarray-based method to identify candidate targets for LY-83583, a suppressor of rapamycin-induced growth inhibition. Finally, Ansari and coworkers used DNA microarrays to interrogate sequence specificity of several fluor-labeled DNA-binding molecules, including engineered polyamide molecules and proteins (10). The cognate site identifier (CSI) microarrays allow rapid and unbiased examination of sequence space and the authors propose using the arrays to determine the sequence preferences of all metazoan DNA-binding proteins and DNA-binding small molecules.
Small-molecule microarrays
The primary use of SMMs to date is ligand discovery. Advantages, including throughput and rapid access to SAR, and disadvantages, including false negatives caused by orientation, have been reviewed previously (26). Typically, a protein of interest is incubated with the array and binding interactions are visualized using a fluorescent readout. Fluorescence intensity is used to rank positive interactions. As most SMM readouts do not correlate directly with affinity, secondary assays, such as thermal-shifts or SPR, are used to confirm positives and to study both the kinetics and the affinity of binding (26). Representative interactions discovered using SMMs are shown in Fig. 1. Interactions of varying affinities that involve proteins from different functional classes, including transcription factors, cytokines, and enzymes, have been discovered. For example, uretupamine and haptamide bind and perturb the functions of two yeast proteins involved in transcriptional regulation and nutrient-sensing (55, 56). Several ligands with differing molecular scaffolds have been identified for calmodulin using SMMs. Calmodioxane (57) and calmoduphilin (58) are products of diversity-oriented syntheses executed by Schreiber and coworkers. NPC-15437, which is a known inhibitor of protein kinase C, binds to calmodulin preferentially when Ca2+ is present in incubation buffer (26). Hsieh-Wilson and coworkers (59) used chondroitin sulfate microarrays to identify chondroitin sulfate-E tetrasaccharide as a ligand for tumor necrosis factor alpha (TNF-α) and demonstrated that the compound disrupted a cytokine-cell-surface receptor interaction. Selective inhibitors of closely related cysteine proteases, cathepsin F and cathepsin K, were identified using the PNA-encoded tetrapeptide acrylate microarrays prepared by Urbina et al. (44). Small-molecule ligands have also been identified for human IgG (25), FKBP12 (60), and Aurora A kinase (61). SMMs have also been used to identify high-affinity interactions between RNA secondary structure motifs and small molecules (62). Microarrays containing drug-like small molecules and carbohydrates have proven very useful for studying carbohydrate-cell interactions because the surface presentation mimics interactions at cell surfaces (22, 63). Enzyme activity assays may also be performed in the SMM format. Yao and coworkers (64) used nanodroplet SMMs for profiling 400 hydroxamate-containing peptide analogs against two matrix metalloproteases and identified selective inhibitors with IC50 values in the nanomolar range. Finally, Dordick and coworkers (65) disclosed a very exciting new application that involves on-array synthesis of natural product analogs using in vitro metabolic pathway construction. The authors demonstrate that these microarrays serve as a platform for synthesis and screening by identifying three inhibitors of Fyn tyrosine kinase.

Figure 1. Representative protein-ligand interactions discovered using small-molecule microarrays. (Adapted from Refs. 26, 27, 44, and 55-62.)
Protein microarrays
The first functional protein and peptide arrays were micropatterned using lithographic methods in 1992 (66). Several years later, MacBeath and Schreiber (13) reported spotted functional protein microarrays capable of detecting protein-protein interactions, protein-ligand interactions, and biochemical activities. Since initial reports, protein microarrays, including peptides, domains, full-length proteins, antibodies, and lysates, have been used in a variety of applications that are reviewed elsewhere (23). MacBeath and coworkers (67) have used protein microarrays to build quantitative protein interaction networks between ErbB receptors with Src homology 2 (SH2) domains and phosphotyrosine binding (PTB) domains. The authors discovered several new binding interactions and proposed that the oncogenic potential of receptor tyrosine kinases may be a function of alterations in binding promiscuity because of changes in EGFR and ErbB2 protein concentrations as both proteins are notably overexpressed in several cancers. The same research group used a similar approach to examine binding of 157 mouse PDZ domains to 217 genome-encoded peptides in an effort to obtain a broad view of selectivity (68). Snyder and coworkers (69) prepared microarrays that contain 282 yeast transcription factors and probed them with fluor-labeled oligonucleotides in an effort to link sequence to binding. Using this approach, the authors defined the binding site of an uncharacterized DNA-binding protein and determined that several of its target genes are involved in stress response and oxidative phosphorylation. The same research group performed biochemical activity analyses on yeast proteome chips to identify in vitro substrates for protein phosphorylation (70). Protein microarrays are also useful tools for target identification (71). As outlined in Fig. 2, Huang et al. (71) prepared biotinylated versions of small molecules known as SMIR3 and SMIR4 that scored as positives in a chemical genetic modifier screen. The labeled compounds were applied to a yeast proteome chip followed by incubation with fluorescently labeled streptavidin. In this fashion, the authors identified lists of candidate protein targets for each compound. One of these proteins, Ybr077cp, was the subject of follow-up studies to confirm the interaction in vivo. Several research groups have used protein and antibody arrays for profiling activities in complex fractions such as whole lysates or proteomic fractions (Fig. 3). Cravatt and coworkers (72) have employed activity-based protein profiling (ABPP) using active-site-directed probes to profile the functional state of enzymes in proteomes. The authors first treated proteome fractions with fluorescent activity-based probes and then applied the fractions to microarrays that contain antibodies against enzymes of interest. Labeled enzymes will be captured so long as a complementary antibody is present. Mahal and coworkers (73) developed a method for analyzing the dynamic glycosylation status of cells by profiling differentially fluor-labeled membrane fractions on microarrays that contain lectin proteins. In analogy to gene-expression profiling, the authors propose this method as a means to make comparisons between glycosylation patterns in different cell states. Finally, reverse-phase protein microarrays, where cell lysate samples are printed and probed with purified antibodies or proteins of interest, have been used to profile cell states and cell types such as the 60 human cancer cell lines (NCI-60) used by the National Cancer Institute to screen small molecules for anticancer activity (Fig. 3b) (74). MacBeath and coworkers (75) used this approach to screen a panel of 84 selected small molecules for their ability to induce different states in the ErbB signaling network. Lysates corresponding to cells treated with small molecules were arrayed and probed with antibodies capable of reporting on the phosphorylation state of selected proteins. This state-based discovery approach may also prove useful in efforts to identify protein targets of bioactive small molecules.
Although less developed relative to microarrays of biomolecules, living microarrays will no doubt play an increasing role in chemical biology. Current applications of this technology include high-throughput and localized transfection (18), high-throughput phenotypic screening of small molecules (52), and loss-of-function screens using RNAi (19, 54). The data gained from these types of experiments should be highly complementary to that gained from the effects of evaluating small molecules using the other microarray platforms. To make the most of the various microarray platforms, standardized array data outputs will be required to address issues that surround reliability and reproducibility of data generated in a microarray format (76). Additionally, databases equipped with Web-based analysis tools will be needed to make the large amount of information generated using microarrays available for public scrutiny (77, 78).

Figure 2. Target identification using yeast proteome microarrays. (a) Using a chemical genetic modifier screen, Huang and coworkers (71) identified small-molecule inhibitors of rapamycin (SMIRs) that suppress the antiproliferative effect of rapamycin in S. cerevisiae. (b) Biotinylated derivatives of SMIR3 and SMIR4 were synthesized for incubation with proteome microarrays in an effort to identify putative protein targets. (c) Binding of biotinylated SMIR3 and SMIR4 to proteins on a yeast proteome chip that contains ~5800 proteins was detected using Cy3-labeled streptavidin. Nine proteins bound to SMIR3 with one protein, Tep1p, annotated as a strong binder. Thirty proteins bound to SMIR4 and four proteins were annotated as strong binders, including Rot1p, YBR077cp, Ras2p, and Met8p.

Figure 3. Changes in protein abundance levels in response to alterations of cell state (e.g., disease or treatment with a small molecule) can be monitored using microarrays. (a) By analogy to transcriptional profiling, antibody microarrays may be used to profile fluorescently labeled proteome fractions. In this scheme, proteins enriched in the compound-treated sample, relative to untreated cells, provide red microarray features. Proteins depleted in the compound-treated sample appear as green features. Proteins that do not change in abundance result in yellow features. Proteins that are absent from the samples, resistant to labeling, or overlabeled such that they no longer recognize the printed antibody give no fluorescent signal. (b) Alternatively, protein fractions from cell lysate samples may be printed on microarrays. Replicate arrays are then probed with different fluor-labeled antibodies of interest to monitor changes in protein abundance. In this scheme, lysate features that contain proteins of interest will be fluorescent (red), whereas lysates that do not contain the protein are not fluorescent (black). Fluorescently labeled control proteins, such as BSA or streptavidin, or antibodies are included as reference points in the arrays (green).
1. Southern EM, Maskos U, Elder JK. Analyzing and comparing nucleic acid sequences by hybridization to arrays of oligonucleotides: evaluation using experimental models. Genomics. 1992; 13:1008-1017.
2. Maskos U, Southern EM. A novel method for the parallel analysis of multiple mutations in multiple samples. Nucleic Acids Res. 1993; 21:2269-2270.
3. Fodor SP, Rava RP, Huang XC, Pease AC, Holmes CP, Adams CL. Multiplexed biochemical assays with biological chips. Nature 1993; 364:555-556.
4. Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 1995; 270:467-470.
5. DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 1997; 278:680-686.
6. Pollack JR, Perou CM, Alizadeh AA, Eisen MB, Pergamenschikov A, Williams CF, Jeffrey SS, Botstein D, Brown PO. Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nat. Genet. 1999; 23:41-46.
7. Kennedy GC, Matsuzaki H, Dong S, Liu WM, Huang J, Liu G, Su X, Cao M, Chen W, Zhang J, Liu W, Yang G, Di X, Ryder T, He Z, Surti U, Phillips MS, Boyce-Jacino MT, Fodor SP, Jones KW. Large-scale genotyping of complex DNA. Nat. Biotechnol. 2003; 21:1233-1237.
8. Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, Volkert TL, Wilson CJ, Bell SP, Young RA. Genome-wide location and function of DNA binding proteins. Science 2000; 290:2306-2309.
9. Bulyk ML, Huang X, Choo Y, Church GM. Exploring the DNA-binding specificities of zinc fingers with DNA microarrays. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:7158-7163.
10. Warren CL, Kratochvil NC, Hauschild KE, Foister S, Brezinski ML, Dervan PB, Phillips GN, Ansari AZ. Defining the sequence-recognition profile of DNA-binding molecules. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:867-872.
11. Ramaswamy S, Golub TR. DNA microarrays in clinical oncology. J. Clin. Oncol. 2002;20:1932-1941.
12. MacBeath G, Koehler AN, Schreiber SL. Printing small molecules onto microarrays and detecting protein-ligand interactions en masse. J. Am. Chem. Soc. 1999; 121:7967-7968.
13. MacBeath G, Schreiber SL. Printing proteins as microarrays for high throughput function determination. Science 2000; 289:1760-1763.
14. Houseman BT, Mrksich M. Carbohydrate arrays for evaluation of protein binding and enzymatic modification. Chem. Biol. 2002; 9:443-454.
15. Park S, Shin I. Fabrication of carbohydrate chips for studying protein-carbohydrate interactions. Angew. Chem. Int. Ed. Engl. 2002; 41:3180-3182.
16. Fang Y, Frutos AG, Lahiri J. Ganglioside microarrays for toxin detection. Langmuir. 2003; 19:1500-1505.
17. Salisbury CM, Maly DJ, Ellman JA. Peptide microarrays for the determination of protease substrate specificity. J. Am. Chem. Soc. 2002; 124:14868-14870.
18. Ziauddin J, Sabatini DM. Microarrays of cells expressing defined cDNAs. Nature 2001; 411:107-110.
19. Bailey SN, Ali SM, Carpenter AE, Higgins CO, Sabatini DM. Microarrays of lentiviruses for gene function screens in immortalized and primary cells. Nat. Methods. 2006; 3:117-122.
20. Nocito A, Kononen J, Kallioniemi OP, Sauter G. Tissue microarrays (TMAs) for high-throughput molecular pathology research. Int. J. Cancer. 2001; 94:1-5.
21. Kanan MW, Rozenman MM, Sakurai K, Snyder TM, Liu DR. Reaction discovery enabled by DNA-templated synthesis and in vitro selection. Nature 2004; 431:545-549.
22. Barrett OJ, Childs JL, Disney MD. Chemical microarrays to identify ligands that bind pathogenic cells. Chem Bio Chem. 2006; 7:1882-1885.
23. Hall DA, Ptacek J, Snyder M. Protein microarray technology. Mech. Ageing Dev. 2007; 128:161-167.
24. Singh-Gasson S, Green RD, Yue Y, Nelson C, Blattner F, Sussman MR, Cerrina F. Maskless fabrication of light-directed oligonucleotide microarrays using a digital micromirror array. Nat. Biotechnol. 1999; 17:974-978.
25. Uttamchandani M, Walsh DP, Yao SQ, Chang YT. Small molecule microarrays: recent advances and applications. Curr. Opin. Chem. Biol. 2005; 9:4-13.
26. Duffner JL, Clemons PA, Koehler AN. A pipeline for ligand discovery using small-molecule microarrays. Curr. Opin. Chem. Biol. 2007; 11:74-82.
27. Uttamchandani M, Walsh DP, Kheronsky SM, Huang X, Yao SQ, Chang YT. Microarrays of tagged combinatorial triazine libraries in the discovery of small-molecule ligands of human IgG. J. Comb. Chem. 2004; 6:862-868.
28. Lee MR, Shin I. Fabrication of chemical microarrays by efficient immobilization of hydrazide-linked substances on epoxide-coated glass surfaces. Angew. Chem. Int. Ed. Engl, 2005; 44:2881-2884.
29. Park S, Shin I. Carbohydrate microarrays for assaying galactosyl-transferase activity. Org. Lett. 2007; 9:1675-1678.
30. Bradner JE, McPherson OM, Mazitschek R, Barnes-Seeman D, Shen JP, Dhaliwal J, Stevenson KE, Duffner JL, Park SB, Neuberg DS, Nghiem P, et al. A robust small-molecule microarray platform for screening cell lysates. Chem. Biol. 2006; 13:493-504.
31. Kanoh N, Asami A, Kawatani M, Honda K, Kumashiro S, Takayama H, Simizu S, Amemiya T, Kondoh Y, Hatakeyama S, Tsuganezawa K, et al. Photo-cross-linked small-molecule microarrays as chemical genomic tools for dissecting protein-ligand interactions. Chem. Asian J. 2006; 1:789-797.
32. Kohn M, Wacker R, Peters C, Schroder H, Soulere L, Breinbauer R, Niemeyer CM, Waldmann H. Site-selective protein immobilization by Staudinger ligation. Angew. Chem. Int. Ed. Engl. 2003; 42:5830-5834.
33. Watzke A, Kohn M, Gutierrez-Rodriguez M, Wacker R, Schroder H, Breinbauer R, Kuhlmann J, Alexandrov K, Niemeyer CM, Goody RS, Waldmann H. Site-selective protein immobilization by Staudinger ligation. Angew. Chem. Int. Ed. Engl. 2006; 45:1408-1412.
34. Schaferling M, Kambhampati D. Protein microarray surface chemistry and coupling schemes. In: Protein Microarray Technology. Kambhampati, D, ed. 2004. Wiley-VCH Verlag GmbH & KGaA, Weinheim. pp. 11-38.
35. Kusnezow W, Jacob A, Walijew A, Diehl F, Hoheisel JD. Antibody microarrays: an evaluation of production parameters. Proteomics 2003; 3:254-264.
36. Shalon D, Smith SJ, Brown PO. A DNA microarray system for analyzing complex DNA samples using two-color fluorescent probe hybridization. Genome Res. 1996; 6:639-645.
37. Angenendt P, Gloker J, Murphy D, Lehrach H, Cahil DJ. Toward optimized antibody microarrays: a comparison of current microarray support materials. Anal. Biochem. 2002; 309:253-260.
38. Stillman BA, Tonkinson JL. FAST slides: a novel surface for microarrays. Biotechniques. 2000; 29:630-635.
39. Wang D, Liu S, Trummer BJ, Deng C, Wang A. Carbohydrate microarrays for the recognition of cross-reactive molecular markers of microbes and host cells. Nat. Biotechnol. 2002; 20:275-281.
40. Fukui S, Feizi T, Galustian C, Lawson AM, Chai W. Oligosaccharide microarrays for high-throughput detection and specificity assignments of carbohydrate-protein interactions. Nat. Biotechnol. 2002; 20:1011-1017.
41. Zhu H, Bilgin M, Bangham R, Hall D, Casamayor A, Bertone P, Lan N, Jansen R, Bidlingmaier S, Houfek T, Mitchell T, Miller P, Dean RA, Gerstein M, Snyder M. Science 2001;293:2101-2105.
42. Lesaicherre ML, Lue RY, Chen GY, Zhu Q, Yao SQ. Intermediated biotinylation of proteins and it’s application in a protein microarray. J. Am. Chem. Soc. 2002; 124:8768-8769.
43. Ko KS, Jaipuri FA, Pohl NL. Fluorous-based carbohydrate microarrays. J. Am. Chem. Soc. 2005; 127:13162-13163.
44. Urbina HD, Debaene F, Jost B, Bole-Feysot C, Mason DE, Kuzmic P, Harris JL, Winssinger N. Self-assembled small- molecule microarrays for protease screening and profiling. ChemBioChem. 2006; 7:1790-1797.
45. Zhu H, Klemic JF, Chang S, Bertone P, Casamayor A, Klemic KG, Smith D, Gerstein M, Reed MA, Snyder M. Analysis of yeast protein kinases using protein chips. Nat. Genet. 2000; 26:283-289.
46. Horiuchi KY, Wang Y, Diamond SL, Ma H. Microarrays for the functional analysis of the chemical-kinase interactome. J. Biomol. Screen. 2006; 11:48-56.
47. Ramachandran N, Larson DN, Stark PR, Hainsworth E, LaBaer J. Emerging tools for real-time label-free detection of interactions on functional protein microarrays. FEBS J. 2005; 272:5412-5425.
48. Kanoh N, Kyo M, Inamori K, Asami A, Nakao A, Osada H. SPR imaging of photo-cross-linked small-molecule arrays on gold. Anal. Chem. 2006; 78:2226-2230.
49. Butcher RA, Schreiber SL. Using genome-wide transcriptional profiling to elucidate small-molecule mechanism. Curr. Opin. Chem. Biol. 2005; 9:25-30.
50. Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, Reich M, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science 2006; 313:1929-1935.
51. Butcher RA, Bhullar BS, Perlstein EO, Marsischky G, LaBaer J, Schreiber SL. Microarray-based method for monitoring yeast overexpression strains reveals small-molecule targets in TOR pathway. Nat. Chem. Biol. 2006; 2:103-109.
52. Bailey SN, Sabatini DM, Stockwell BR, Microarrays of small molecules embedded in biodegradable polymers for use in mammalian cell-based screens. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:16144-16149.
53. Disney MD, Magnet S, Blanchard JS, Seeberger PH. Aminoglycoside microarrays to study antiobiotic resistance. Angew. Chem. Int. Ed. Engl. 2004; 43:1591-1594.
54. Wheeler DB, Bailey SN, Guertin DA, Carpenter AE, Higgins CO, Sabatini DM. RNAi living-cell microarrays for loss-of-function screens in Drosophila melanogaster cells. Nat. Methods. 2004; 1:103-104.
55. Kuruvilla FG, Shamji AF, Sternson SM, Hergenrother PJ, Schreiber SL. Dissecting glucose signaling with diversity-oriented synthesis and small-molecule microarrays. Nature 2002; 416:653-657.
56. Koehler AN, Shamji AF, Schreiber SL. Discovery of an inhibitor of a transcription factor using small molecule microarrays and diversity-oriented synthesis. J. Am. Chem. Soc. 2003; 125:8420- 8421.
57. Wong JC, Sternson SM, Louca JB, Hong R, Schreiber SL. Modular synthesis and preliminary biological evaluation of stereochemically diverse 1,3-dioxanes. Chem. Biol. 2004; 11:1279-1291.
58. Barnes-Seeman D, Park SB, Koehler AN, Schreiber SL. Expanding the functional group compatibility of small-molecule microarrays: discovery of novel calmodulin ligands. Angew. Chem. Int. Ed. Engl. 2003; 42:2376-2379.
59. Tully SE, Rawat M, Hsieh-Wilson LC. Discovery of a TNF-alpha antagonist using chondroitin sulfate microarrays. J. Am. Chem. Soc. 2006;128:7740-7741.
60. Clemons PA, Koehler AN, Wagner BK, Sprigings TG, Spring DR, King RW, Schreiber SL, Foley MA. A one-bead, one-stock solution approach to chemical genetics: part 2. Chem. Biol. 2001; 8:1183-1195.
61. Miao H, Tallarico JA, Hayakawa H, Munger K, Duffner JL, Koehler AN, Schreiber SL, Lewis TA. Ring-opening and ringclosing reactions of a shikimic acid-derived substrate leading to diverse small molecules. J. Comb. Chem. 2007; 9:245-253.
62. Kwon SK, Lee MY, Ku B, Sherman DH, Dordick JS. High-throughput, microarray-based synthesis of natural product analogues via in vitro metabolic pathway construction. ACS Chem. Biol. 2007; 2:419-425.
63. Disney MD, Childs-Disney JL. Using selection to identify and chemical microarray to study the RNA internal loops recognized by 6’-N-acylated kanamycin A. ChemBioChem. 2007; 8:649-656.
64. Disney MD, Seeberger PH. The use of carbohydrate microarrays to study carbohydrate-cell interactions and to detect pathogens. Chem. Biol. 2004; 11:1701-1707.
65. Wang J, Uttamchandani M, Sun LP, Yao SQ. Activity-based high-throughput profiling of metalloprotease inhibitors using small molecule microarrays. Chem. Commun. (Camb). 2006; (7):717- 719.
66. Britland S, Perez-Arnuad E, Clark P, McGinn B, Connolly P, Moores G. Micropatterning proteins and synthetic peptides on solid supports: a novel application for microelectronics fabrication technology. Biotechnol. Prog. 1992; 8:155-160.
67. Jones RB, Gordus A, Krall JA, MacBeath G. A quantitative protein interaction network for the ErbB receptors using protein microarrays. Nature 2006; 439:168-174.
68. Stiffler MA, Chen JR, Grantcharova VP, Lei Y, Fuchs D, Allen JE, Zaslavakaia LA, MacBeath G. PDZ domain binding selectivity is optimized across the mouse proteome. Science 2007; 317:364-369.
69. Ho SW, Jona G, Chen CT, Johnston M, Snyder M. Linking DNA-binding proteins to their recognition sequences by using protein microarrays. Proc. Natl. Acad. Sci.U.S.A. 2006; 103:9940- 9945.
70. Ptacek J, Devgan G, Michaud G, Zhu H, Zhu X, Fasolo J, Guo H, Jona G, Breitkreutz A, Sopko R, et al. Global analysis of protein phosphorylation in yeast. Nature 2005; 438:679-684.
71. Huang J, Zhu H, Haggarty SJ, Spring DR, Hwang H, Jin F, Snyder M, Schreiber SL. Finding new components of the target of rapamycin (TOR) signaling network through chemical genetics and proteome chips. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:16594-16599.
72. Sieber SA, Mondala TS, Head SR, Cravatt BF. Microarray platform for profiling enzyme activities in complex proteomes. J. Am. Chem. Soc. 2004; 126:15640-15641.
73. Pilobello KT, Slawek DE, Mahal LK. A ratiometric lectin microarray approach to analysis of the dynamic mammalian glycome. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:11534-11539.
74. Nishizuka S, Charboneau L, Young L, Major S, Reinhold WC, Waltham M, Kouros-Mehr H, Bussey KJ, Lee JK, Espina V, Munson PJ, Petricoin E, Liotta LA, Weinstein JN. Proteomic profiling of the NCI-60 cancer cell lines using new high-density reverse-phase lysate microarrays. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:14229-14234.
75. Sevecka M, MacBeath G. State-based discovery: a multidimensional screen for small-molecule modulators of EGF signaling. Nat. Methods. 2006; 3:825-831.
76. MAQC Consortium. The Microarray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat. Biotechnol. 2006; 24:1151-1161.
77. Demeter J, Beauheim C, Gollub J, Hernandez-Boussard T, Jin H, Maier D, Matese JC, Nitzberg M, Wymore F, Zachariah ZK, Brown PO, Sherlock G, Ball CA. The Stanford Microarray Database: implementation of new analysis tools and open source release of software. Nucleic Acids Res. 2007; 35:D766-D770.
78. Tolliday N, Clemons PA, Ferraiolo P, Koehler AN, Lewis TA, Li X, Schreiber SL, Gerhard DS, Eliasof S. Small molecules, big players: the National Cancer Institute’s Initiative for Chemical Genetics. Cancer Res. 2006; 66:8935-8942.
Further Reading
Butcher RA, Schreiber SL. A microarray-based protocol for monitoring the growth of yeast overexpression strains. Nat. Protoc. 2006; 1:569-576.
Bradner JE, McPherson OM, Koehler AN. A method for the covalent capture and screening of diverse small molecules in a microarray format. Nat. Protoc. 2006; 1:2344-2352.
Chattopadhaya S, Tan LP, Yao SQ. Strategies for site-specific protein biotinylation using in vitro, in vivo, and cell-free systems; toward functional protein arrays. Nat. Protoc. 2006; 1:2386-2398.
Erlich JR, Qin S, Liu BC. The ‘reverse capture’ autoantibody microarray: a native antigen-based platform for autoantibody profiling. Nat. Protoc. 2006; 1:452-460.
Fiegler H, Redon R, Carter NP. Construction and use of spotted large-insert clone DNA microarrays for the detection of genomic copy number changes. Nat. Protoc. 2007; 2:577-587.
Hill HD, Mirkin CA. The biobarcode assay for the detection of protein and nucleic acid targets using DTT-induced ligand exchange. Nat. Protoc. 2006; 1:324-336.
Hsu KL, Mahal LK. A lectin microarray approach for the rapid analysis of bacterial glycans. Nat. Protoc. 2006; 1:543-549.
Jing R, Bolshakov V, Flavell AJ. The tagged microarray marker (TAM) method for high-throughput detection of single nucleotide and indel polymorphisms. Nat. Protoc. 2007; 2:168-177.
Keene JD, Komisarow JM, Friedersdorf MB. RIP-chip: the isolation and identification of mRNAs, microRNAs and protein components of the ribonucleoprotein complexes from cell extracts. Nat. Protoc. 2006; 1:302-307.
Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat. Protoc. 2006; 1:729-748.
Liu B, Bazan GC. Synthesis of cationic conjugated polymers for use in label-free DNA microarrays. Nat. Protoc. 2006; 1:1698-1702.
Chembank. http://chembank.broad.harvard.edu/.
National Human Genome Research Institute (NHGRI) Microarray Project. http://research.nhgri.nih.gov/microarray/.
Protein Microarrays. http://www.stanford.edu/btomooka/index.htm.
Stanford Microarray Database. http://genome-www5.stanford.edu/.
See Also
Forward Chemical Genetics
Array-Based Techniques for Glycans: Development and Applications
Peptide Combinatorial Libraries
Transcript Profiling
Receptor-Ligand Interactions
Combinatorial Libraries: Overview of Applications in Chemical Biology