CHEMICAL BIOLOGY
Cytosolic Glycosylated Proteins, Chemistry of
Clifford A. Toleman and Jeffrey E. Kudlow, University of Alabama at Birmingham, Alabama
doi: 10.1002/9780470048672.wecb117
O-GlcNAcylation is a prevalent posttranslational modification that occurs when a molecule of the monosaccharide N-acetylglucosamine (GlcNAc) is adjoined to a serine or a threonine residue of a cellular protein. This modification can influence a protein in many of the same ways that a serine or threonine phosphorylation can. In addition, much like the kinases and phosphatases involved in phosphorylation/dephosphorylation, unique enzymes are responsible for the addition and removal of O-GlcNAc modifications, with an O-GlcNAc transferase that catalyzes the addition and an O-GlcNAcase that is responsible for the removal. This review will give a summate description of the diverse biochemical roles that this O-GlcNAcylation can play on individual proteins, and on whole animal systems, and will detail the current understanding of the enzymes involved in its maintenance. Finally, we discuss here the recent advances that have been made in the development of tools to study O-GlcNAcylation.
O-GlcNAcylation, which was first described just over 20 years ago, is a ubiquitous posttranslational modification, whereas a sugar molecule, N-acetylglucosamine (GlcNAc), is added to precise serine or threonine residues on cellular proteins. This modification can alter the physical properties of a protein in several ways, and it is highly analogous to posttranslational phosphorylation. The study of, and the understanding of, phosphorylation is well established, and in recent years, it has resulted in many advances in the comprehension of several proteins that are O-GlcNAc modified, as well as how this modification influences individual protein activities. The last five years have also brought about a wealth of biochemical information that concerns the enzymes accountable for the regulation of this modification, the O-GlcNAc transferase and the
O-GlcNAcase, which includes their domain structures and the characterization of their catalytic activities. The mapping of O -GlcNAcylation sites has also experienced highly significant progress. Although O-GlcNAcylation still lags behind phosphorylation in popularity, this profusion of information is leading to the firm establishment of the importance of O-GlcNAc in cell and molecular biology.
Biologic Background
When glucose enters a cell, its primary fate is to be phosphorylated and converted to glucose-6-phosphate. The glucose-6-phosphate is then converted to fructose-6-phosphate, whereby it awaits one of several fates. The greater percentage will eventually be converted either to glycogen for storage in skeletal muscle and in liver or broken down for energy in the glycolytic pathway. Approximately 2-5% of the fructose-6-phosphate, however, enters the hexosamine biosynthetic pathway (HBP), where it ultimately will be converted to the high energy compound UDP-N-acetylglucosamine (UDP-GlcNAc). These sugar residues are linked commonly to proteins in two different ways. N-linked glycosylation occurs when the attachment is made to the amide nitrogen of asparagine side chains. N-linked glycosylations contain a minimum of four sugars in addition to the terminal GlcNAc, and this modification typically is implicated in the directing of proteins through the endoplasmic reticulum-Golgi-plasmalemma pathway. The second fate of the UDP-GlcNAc is its O -linkage to proteins (Fig. 1).
O-GlcNAc as a posttranslational modification
First described in 1984, O-linked GlcNAcylation is a form of posttranslational modification where a single sugar, the monosaccharide GlcNAc, is affixed via a beta linkage to the hydroxyl group of specific serine and threonine residues on target proteins (1). In rare cases, O-linked N-acetylglucosamine (O-GlcNAc) linkage can occur on lysine side chains. Although this modification has not been witnessed in bacteria, it is otherwise ubiquitous in all eukaryotic cells that have been studied. O-GlcNAcylation can occur on both extracellular and intracellular proteins and can be found on both cytosolic and nuclear members of the latter (1). O-GlcNAc additions are reversible, highly dynamic, and occur on proteins involved in most, if not all, processes of fundamental importance to a cell, including transcription, DNA replication, protein-folding, cytoskeletal regulation, translation, metabolic and signaling pathways, nuclear import-export, protein degradation, and vesicular trafficking, among many others. In addition to these downstream roles, the HBP is speculated to act as a global cell nutrient sensor. Given the makeup of the substrate UDP-GlcNAc, its levels are governed by the availability and the metabolism of amino acids (glutamine is used as the amine donor), fatty acids (which supply the eventual acetate), nucleotides (for UDP), and glucose in particular (for the sugar backbone). The global accumulation of O-GlcNAcylated proteins, as a result of an increase in glucose uptake and therefore UDP-GlcNAc availability, may act as negative feedback, directly or indirectly, to downregulate proteins involved in additional glucose import (2, 3). This may be the case with catabolism and anabolism of the other constituents of UDP-GlcNAc as well.
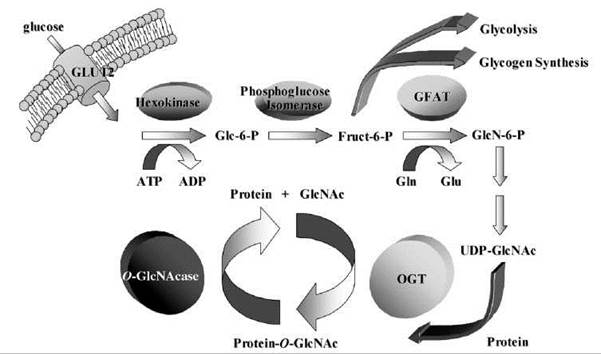
Figure 1. When entry into a cell occurs, glucose is converted to glucose-6-phosphate (Glc-6-P) and then into fructose-6-phosphate (Fruct-6-P). Approximately 5% of the fructose-6-phosphate enters the hexosamine pathway, where the enzyme GFAT converts it to glucosamine-6-phosphate (GlcN-6-P), in a process that requires glutamine's (Gln) conversion to glutamate (Glu). Eventually, the glucosamine-6-phosphate is converted to UDP-GlcNAc, and this can be added to protein serines or threonines by the enzyme O-GlcNAc transferase (OGT). O-GlcNAcase is the enzyme that removes this modification.
O-GlcNAcylation in disease states
As a result of all of the above, the deregulation of protein-O-GlcNAc homeostasis has been linked to several disease pathologies, particularly diabetes, cancer, and Alzheimer’s disease. For example, increases in protein-O-GlcNAc—either by increasing the flux through the HBP, an overexpressing the enzyme responsible for its addition to proteins (P-N-acetylglucosaminyl transferase or OGT), or inhibiting the enzyme responsible for its removal (O-GlcNAcase)—can recapitulate several hallmark features of type II diabetes (OGT and O-GlcNAcase will be discussed). In fat or muscle tissue, increased O-GlcNAc leads to insulin resistance and to the hyperleptinemia associated with increased fat synthesis and storage (4). In liver cells, an increase in O-GlcNAc results in an upregulation of glycogen storage, impaired glucose tolerance, and whole animal hyperlipidemia and obesity (5). In pancreatic P-cells, inflations in O-GlcNAc produce apoptosis (6), hyperinsulinemia, and insulin resistance (7); this inflation is postulated to be primarily because many proteins involved in the insulin-signaling pathways are O-GlcNAced, including IRS-1, Akt, and PI-3 kinase. Finally, the O-GlcNAcase gene is located in a characterized diabetes susceptibility locus, and a naturally occurring single nucleotide polymorphism discovered within this gene was found to correlate with an increase in diabetic risk (8).
Second, because several proteins have been demonstrated to be O-GlcNAced in a cell-cycle-dependent manner, and because cellular O-GlcNAc levels are highly correlative to the cell cycle (9), as well as in response to mitogens, growth factors, and cell stresses [see review by Zachara and Hart (10)], the disruption of O-GlcNAc pathways can be tumorigenic. Altering flux through the hexosamine pathway, providing more O-GlcNAc substrate for target proteins, has been shown to modify the growth rate of various cell types (11). In addition, overexpression of OGT, which results in more protein-O-GlcNAcylation, can result in polyploidy and defects in cytokinesis. Overexpression of the converse enzyme, O-GlcNAcase, influences cell cycle, which results in mitotic exit delays, altered cyclin expression, nuclear morphological differences, and delayed mitotic phosphorylation (9). Thus, cancerous phenotypes can result from faulty O-GlcNAcylation.
Finally, much evidence indicates that O-GlcNAc signaling is involved in neurodegenerative diseases. The potential role of O-GlcNAc in the pathogenesis of Alzheimer’s disease, in particular, has drawn much attention. The OGT and O-GlcNAcase enzymes are highly enriched in the brain and are particularly high in the Purkinje cells. The pathogenesis of Alzheimer’s disease involves, among other events, the accumulation of P-amyloid and tau aggregates, both of which have been shown to be O-GlcNAc modified and increasingly are modified in Alzheimer’s stricken cells (12). Their ability to be degraded and cleared by the proteosomes in hippocampal cells is inhibited by increased O-GlcNAc levels (13, 14). In addition to being a hot spot for diabetes susceptibility genes, the chromosomal locus for the O-GlcNAcase gene is a candidate locus for late-onset Alzheimer’s disease. Interestingly, the locus for the single OGT gene is very near the locus responsible for dystonia-Parkinsonism syndrome.
The Roles of an O-GlcNAc Modification
Although the explication of O-GlcNAc as a posttranslational modification has taken time (it has been just over 20 years since its discovery), the rate of its appreciation is similar to that of phosphorylation after its discovery. Hundreds of proteins have been demonstrated to be O-GlcNAc modified (Table 1), with a great many more as yet untested that possess potential O-GlcNAcylation sites. The roles that an O-GlcNAc modification can play on a protein have been found to be comparable to those of O -phosphorylation. Although GlcNAc is a neutral molecule, and does not possess the strong negative charge of a phosphate, its possession of numerous negatively polar hydroxyl groups allows it to perform many of the same reversible functions.
The addition of O-GlcNAc can alter protein-protein interactions. For example, adding it to the ubiquitous transcription factor Sp1 can inhibit its binding to TAFII110 and can prevent gene transcription (15), and its addition to the chaperone HSP60 can obstruct HSP60-Bax interaction, which results in Bax translocation to the mitochondria in initiating steps of the apoptotic response (16). Conversely, O-GlcNAc modification on the signal transducer Stat5a can enhance the interaction of Stat5a with the transcriptional coactivator CBP (17). Protein-DNA interactions can also be influenced by an O-GlcNAc addition or loss, as is observed, for example, on the PDX-1 transcription factor, where increased O-GlcNAcylation on PDX-1 increases its DNA binding affinity (18). Within the transcription factor Oct-2, O-GlcNAcylation likewise regulates DNA-binding specificity (19).
O-GlcNAc modifications can also lead to the stabilization of a target molecule. O-GlcNAcylation of plakoglobin, which is a component of adherens junctions and desmosomes, increases the stability of the protein and thereby promotes cell-cell adhesion (20). Another example occurs with eIF2-p67 complexes, where an O-GlcNAcylation event on p67 leads to an increased half-life of these components, which allows the subsequent activation of the eIF2 a subunit required for protein translation (21). Furthermore, O-GlcNAc can contribute directly to global protein stability by modifying the ATPase subunit Rpt2 within proteasome caps, which results in the inhibition of proteasomal degradation (14).
Like phosphorylation, O-GlcNAcylation can activate or inactivate a protein through conformational changes. It is suggested that O-GlcNAc modifications on Stat5 are involved in its transient activation (22), and its addition to P3 integrin is expected to increase the protein’s outside-in signaling necessary for cytoskeletal rearrangements and cell spreading (23). Conversely, O-GlcNAc modification can inhibit the signaling activity of PLC-P1, which is an enzyme involved in the generation of important second messengers that trigger intracellular Ca2+ release (24) or can result in the inactivation of the nitric oxide synthase eNOS (25).
Along with the above phenomena, the addition of O-GlcNAc influences a target protein’s cellular localization. According to several studies, it seems that O-GlcNAc may act as a nuclear localization signal. O-GlcNAcylated proteins seem to be more prominent in nuclear over cytosolic fractions (26), and proteins such as Akt1, mTOR α4, and Sp1 translocate to the nucleus after O-GlcNAc modification. Exceptions exist, as witnessed with a viral protein vJun, where O-GlcNAc added near the NLS influences nuclear import (27) negatively. The nuclear pore is considerably O-GlcNAc modified, and this can influence the transport into or out of the nucleus. The direct influence of O-GlcNAc on pore components is still being explored.
Finally, because this modification occurs on serine and threonine residues, it can also compete directly with an O-phosphory lation event at an identical or nearby site to provide an additional level of regulation on a specific protein. To date, all described O-GlcNAc-modified proteins have been found to be phosphoproteins (28, 29, 30). Such competition between O-GlcNAc and O-phosphate binding occurs with many examples listed above as well as with others.
Table 1. Current listing of identified OGlcNAc modified proteins

The Enzymes that Regulate O-GlcNAcylation
O-GlcNAc transferase
The enzyme that catalyzes the reaction between the target protein and the UDP-GlcNAc, with UDP being the leaving group, is OGT. OGT is a highly conserved, 1036-amino-acid protein found in all metazoans, and it is expressed ubiquitously in all tissues, although mRNA and protein levels can vary. OGT can be found in both the nucleus and the cytosol; however, localization studies have revealed a slight preference for the nucleus (31). The OGT protein is encoded by a single, highly conserved gene mapped to a location on human chromosome Xq13.1. Murine as well as cell knockouts have proven to be lethal, which indicates that the ability to glycosylate proteins is essential to complete embryogenesis and overall individual cell viability (32, 33). Conversely, overexpression of OGT is also toxic to cells, which suggests that, in addition to the necessity of the presence of O-GlcNAc modification, the maintenance of proper protein-O-GlcNAc levels is also essential for cellular survival. O-GlcNAc transfer is mediated by two C-terminal catalytic domains (CD I and CD II) that form a pocket together with both UDP-GlcNAc binding sites and a catalytic center (34, 35). In addition to its catalytic domains, OGT contains 11.5 tetratricopeptide (TPR) motifs in the protein’s N-terminus. TPR motifs are 34-amino-acid structures with several electrostatic surfaces that function primarily in protein-protein interactions, and this allows OGT to interact with a large number of proteins and with itself (36). OGT has at least three known isoforms with different cellular localizations (37). These isoforms all contain the catalytic domains; however, they differ in their number of TPR motifs, which suggests that the variations in cellular localization are in part caused by the targeting of its binding partners (37).
Because OGT has several binding partners, substrates, and varying expression levels and localization, it is not surprising that it is regulated tightly. To date, OGT has been shown to be O-GlcNAc modified as well as tyrosine phosphorylated (35, 38). The roles and sites of these modifications have not yet been elucidated, but studies suggest the tyrosine phosphorylation may have an activating effect on the enzyme’s catalysis (39). OGT substrate specificity and localization are regulated by its binding partners as well as by its own multimerization (35). Finally, OGT activity can also be influenced by feedback mechanisms. OGT is highly responsive to intracellular UDP-GlcNAc levels (35), and free UDP is a potent inhibitor of this substrate recognition (35). OGT catalytic activity has also been demonstrated to be inhibitable by the drug compound alloxan, which is a uracil analog (40). Naturally, neither of these inhibitors is specific for OGT activity within a cell. Currently, the development of OGT-specific inhibitors has only been preliminarily characterized (41).
O-GlcNAcase
β-O-linked N-acetylglucosamidase, or O-GlcNAcase, is a 917-amino-acid enzyme responsible to remove O-GlcNAc modifications on a protein selectively. Like OGT, O -GlcNAcase can be found in the nucleus and in the cytosol of all tissues and organisms higher than yeast. Also, like OGT, O-GlcNAcase is expressed by a single gene product at a chromosomal locus at 10 q24.1-.3 (in humans) that encodes at least three alternative splice variants in addition to the full-length product. The O-GlcNAcase active site, a TIMα/β8 barrel (42, 43), resides in the protein’s N-terminus (44, 45) and is part of a larger enzyme that contains an acetyltransferase (AT) domain in the protein’s C-terminus. O-GlcNAcase has been shown to have acetyltransferase activity, at least for histone substrates, and therefore, it is sometimes referred to as NCOAT (nuclear-cytoplasmic O-GlcNAcase and acetyltransferase) to reflect both activities (46). Two naturally occurring splice variants of this enzyme lack a portion of the O-GlcNAcase active site (exons 8 and 9, encoding amino acids 250-345 and 250-398, respectively), and therefore, they are defunct for O-GlcNAcase activity (46). The third variant lacks the AT domain (47) completely. O-GlcNAcase/NCOAT has been shown to be a substrate of the pro-apoptotic protease caspase-3. The cleavage site flanks the C-terminus of the O-GlcNAcase domain, and because the cleaved products retain their respective activities (46, 48), this may regulate/deregulate these activities as well as disconnect their locality within a cell (48).
Although it is speculated that O-GlcNAcase activity must be regulated highly, little progress has been made to reveal posttranslational modifications that may occur on the protein. O-GlcNAcase has been shown to be O-GlcNAc modified, although it has not been elucidated as to whether this would have an activating or inhibiting effect on the enzyme (37). Like with OGT, O-GlcNAc modification on O-GlcNAcase may represent a unique feedback mechanism to help regulate cellular protein O-GlcNAc levels by regulating the enzymes responsible for the modification themselves. In addition to the variants and caspase cleavage products mentioned above, it is interesting that O-GlcNAcase exhibits strong direct interaction with OGT. It does so via a domain in the middle of the enzyme and through the N-terminus and first six TPRs of OGT (49). To prevent a futile cycle of glycosylation/deglycosylation that surrounds these complexes, the regulation of these enzymes becomes of paramount importance.
Several O-GlcNAcase inhibitors have been described, the most popular are streptozotocin (STZ) and PUGNAc. Both of these compounds are substrate mimetics that inhibit O-GlcNAcase activity by resembling the natural substrate’s oxazoline transition state after its entrance into the active site (50, 51). Unfortunately, the above inhibitors are nonselective and can inhibit other glycosyl hydrolases; therefore, they are a detriment to a multitude of cell pathways. Another, more potent transition state analog, N-acetylglucosamine-thiazoline (NAG-thiazoline), has been described recently (5). The increased potency likely is because the compound already resembles the oxazoline intermediate before it is exposed to the enzyme. Macauley et al. (52) have began recently to generate NAG-thiazoline derivatives with augmentations to the thiazoline ring. In particular, the addition of a butyl chain to the ring caused a dramatic increase in potency over the NAG-thiazoline parent compound and exhibited a selectivity toward O-GlcNAcase over lysosomal hexosaminidases. In addition, Stubbs et al. (53) and Kim et al. (54) have begun to characterize PUGNAc derivatives. These analogs involve alkyl extensions, and whereas the extensions are the same and in a position analogous to those of the NAG-thiazoline derivatives, the extensions are somewhat less potent and specific for O-GlcNAcase, although their usefulness may yet be evident in different contexts. The increased potency and selectivity of these compounds over their precursors represent important advances and will make them invaluable tools in the study of O-GlcNAcylation. Furthermore, Kim et al. (55) have exploited this butyl-extension to increase the sensitivity of a fluorogenic O-GlcNAcase substrate, giving the field a promising high-throughput imaging tool for the analysis of O-GlcNAc functions.
Emerging approaches for the Study of O-GlcNAcylation
Until the last decade, proficient methods to study O-GlcNAcylation on a target substrate had been slow to develop. In whole-cell systems, the use of OGT or O-GlcNAcase inhibitors—or the overexpression of these proteins—is toxic or affects too many cellular pathways to interpret results directly. In isolatable systems, recombinant OGT or O-GlcNAcase, together with their respective inhibitors, can be used to identify target substrates, either by radiolabeling using UDP-(3H)GlcNAc or by Western blotting using protein-O-GlcNAc specific antibodies such as RL2 or CTD110.6. In several cases, the subsequent effect of the O-GlcNAc modification on the protein’s function could be determined in isolatable in vitro assays. More commonly, the effect of the O-GlcNAc modification on the protein was never fully understood, because the sites of O-Glc NAcylation could not be detected. This limitation prevents mutational analysis, and the direct investigation of the role of the O-GlcNAc in purified assays, in signaling pathways and in whole-cell and animal contexts.
Traditional methods to map posttranslational modification sites, like those of phosphorylation, have been anchored by protein digest and mass spectroscopic (MS) approaches (for a review on the classic evaluation and for MS analyses of O-glycans, see Reference (56)). Unfortunately, like many posttranslational modifications, O-GlcNAcylation occurs routinely on a protein population with substoichiometric frequency, which results in a very small detectable population of a O-GlcNAc-modified product. Also, much like O-phosphate additions, the protein-O-GlcNAc bond is labile and is detached by collision-induced dissociation (CID) during MS analysis. Often, the bond is lost before it can be detected on the peptides analyzed (57, 58). Phosphate modifications, however, can overcome this limitation by enriching the peptide mixtures through chromatographic approaches, for example, by using immobilized metal ion affinity columns. Such methods to enrich O-GlcNAc-modified peptides specifically are beginning to be optimized.
Recent advances have been developed to enrich O-GlcNAcylated peptides by using a lectin weak affinity chromatography (LWAC) approach. In the LWAC approach, a column of wheat germ agglutinin (WGA), which has binding affinity for GlcNAc as well as sialic acid, is used to separate and enrich GlcNAcylated peptides from the non-GlcNAcylated population (57). Although the binding affinity is weak, with dissociation constants in the 10-millimolar range, the interaction is sufficient to retard the rate of flow of GlcNAcylated peptides by HPLC enough to separate them for MS analysis and for sequencing. It has also been shown that such WGA chromatography is sufficient enough to purify proteins for 2-D gel electrophoresis and for subsequent MS proteomic analyses (59). These protocols can be used both on purified protein digests as well as in cellular extracts, and they represent a key advance in identifying O-GlcNAc modification sites.
A second promising method for mapping of O-GlcNAcylation sites involves a P-elimination/Michael addition with the DTT (BEMAD) approach (57, 58). This technique can be used alone, or in conjunction with, the above LWAC method. Briefly, the use of a strong base such as sodium hydroxide can result in the P-elimination of the C-CH2-O-GlcNAc to C=CH2, where the first C is within the protein backbone on the digested peptides. This technique is followed by a Michael addition of DTT to the CH2 group (other “tags” can be added through this technique as well). The result is a formerly O -GlcNAced site modified so that it can be enriched by chromatography (and is less labile), as mentioned above for O -phosphate peptide enrichment. In the case of BEMAD, an activated thiol-Sepharose can purify the DTT-tagged peptides, although a variety of columns can be used depending on the tag chosen. These affinity-tag methods have advanced enough so that they can be used to label O -GlcNAc sites selectively, whereas other P-elimination susceptible sites, like those of O-phosphate, are left undisturbed.
Affinity tags have also been developed to circumvent the P-elimination. The first uses an engineered galactosyltransferase to apply a UDP-ketone analog in the place of O-GlcNAc, which can be biotinylated for purification (60, 61). This strategy has been implemented successfully in both pure protein preparations and whole cell lysates. The second strategy exploits a phenomenon first reported by Vocadlo et al., in which N -azidoacetylglucosamine can be used as a substrate for both OGT as well as O-GlcNAcase (62). This method can be applied to cell culture, where a peracetylated GlcNAc azide analog is incubated with the cells (along with an O-GlcNAcase inhibitor to prevent deglycosylation). After the incorporation of the analog at O-GlcNAc sites, the mixture can be treated with a biotinylated phosphine reagent to tag the sites of incorporation for streptavidin purification (63).
References
1. Torres CR, Hart GW. Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence for O-linked GlcNAc. J. Biol. Chem. 1984; 259:3308-3317.
2. Wells L, Vosseller K, Hart GW. A role for N-acetylglucosamine as a nutrient sensor and mediator of insulin resistance. Cell. Mol. Life Sci. 2003; 60:222-228.
3. Vosseller K, Wells L, Lane MD, Hart GW. Elevated nucleocytoplasmic glycosylation by O-GlcNAc results in insulin resistance associated with defects in Akt activation in 3T3-L1 adipocytes. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:5313-5318.
4. McClain DA, Lubas WA, Cooksey RC, Hazel M, Parker GJ, Love DC, Hanover JA. Altered glycan-dependent signaling induces insulin resistance and hyperleptinemia. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:10695-10699.
5. Veerababu G, Tang J, Hoffman RT, Daniels MC, Hebert LF Jr, Crook ED, Cooksey RC, McClain DA. Overexpression of glutamine: fructose-6-phosphate amidotransferase in the liver of transgenic mice results in enhanced glycogen storage, hyperlipidemia, obesity, and impaired glucose tolerance. Diabetes 2000; 49:2070-2078.
6. Liu K, Paterson AJ, Chin E, Kudlow JE. Glucose stimulates protein modification by O-linked GlcNAc in pancreatic beta cells: Linkage of O-linked GlcNAc to beta cell death. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:2820-2825.
7. Tang J, Neidigh JL, Cooksey RC, McClain DA. Transgenic mice with increased hexosamine flux specifically targeted to beta-cells exhibit hyperinsulinemia and peripheral insulin resistance. Diabetes 2000; 49:1492-1499.
8. Lehman DM, Fu DJ, Freeman AB, Hunt KJ, Leach RJ, Johnson-Pais T, Hamlington J, Dyer TD, Arya R, Abboud H, Goring HH, Duggirala R, Blangero J, Konrad RJ, Stern MP. A single nucleotide polymorphism in MGEA5 encoding O-GlcNAc-selective N-acetyl-beta-D glucosaminidase is associated with type 2 diabetes in Mexican Americans. Diabetes 2005; 54:1214-1221.
9. Slawson C, Zachara NE, Vosseller K, Cheung WD, Lane MD, Hart GW. Perturbations in O-linked beta-N-acetylglucosamine protein modification cause severe defects in mitotic progression and cytokinesis. J. Biol. Chem. 2005; 280:32944-32956.
10. Zachara NE, Hart GW. O-GlcNAc a sensor of cellular state: the role of nucleocytoplasmic glycosylation in modulating cellular function in response to nutrition and stress. Biochim. Biophys. Acta 2004; 1673:13-28.
11. Bekesi JG, Winzler RJ. Inhibitory effects of D-glucosamine on the growth of Walker 256 carcinosarcoma and on protein, RNA, and DNA synthesis. Cancer Res. 1970; 30:2905-2912.
12. Griffith LS, Schmitz B. O-linked N-acetylglucosamine is upregulated in Alzheimer brains, Biochem. Biophys. Res. Commun. 1995; 213:424-431.
13. Liu K, Paterson AJ, Zhang F, McAndrew J, Fukuchi K, Wyss JM, Peng L, Hu Y, Kudlow JE. Accumulation of protein O-GlcNAc modification inhibits proteasomes in the brain and coincides with neuronal apoptosis in brain areas with high O-GlcNAc metabolism. J. Neurochem. 2004; 89:1044-1055.
14. Zhang F, Su K, Yang X, Bowe DB, Paterson AJ, Kudlow JE. O-GlcNAc modification is an endogenous inhibitor of the proteasome. Cell 2003; 115:715-725.
15. Yang X, Su K, Roos MD, Chang Q, Paterson AJ, Kudlow JE. O-linkage of N-acetylglucosamine to Sp1 activation domain inhibits its transcriptional capability. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:6611-6616.
16. Kim HS, Kim EM, Lee J, Yang WH, Park TY, Kim YM, Cho JW. Heat shock protein 60 modified with O-linked N-acetylglucosamine is involved in pancreatic beta-cell death under hyperglycemic conditions. FEBS Lett. 2006; 580:2311-2316.
17. Gewinner C, Hart G, Zachara N, Cole R, Beisenherz-Huss C, Groner B. The coactivator of transcription CREB-binding protein interacts preferentially with the glycosylated form of Stat5. J. Biol. Chem. 2004; 279:3563-3572.
18. Gao Y, Miyazaki J, Hart GW. The transcription factor PDX-1 is post-translationally modified by O-linked N-acetylglucosamine and this modification is correlated with its DNA binding activity and insulin secretion in min6 beta-cells. Arch. Biochem. Biophys. 2003; 415:155-163.
19. Ahmad I, Hoessli DC, Walker-Nasir E, Rafik SM, Shakoori AR, Nasir ud D. Oct-2 DNA binding transcription factor: functional consequences of phosphorylation and glycosylation. Nucleic Acids Res. 2006; 34:175-184.
20. Hu P, Berkowitz P, Madden VJ, Rubenstein DS. Stabilization of plakoglobin and enhanced keratinocyte cell-cell adhesion by intracellular O-glycosylation. J. Biol. Chem. 2006; 281:12786-12791.
21. Datta R, Choudhury P, Ghosh A, Datta B. A glycosylation site, 60SGTS63, of p67 is required for its ability to regulate the phosphorylation and activity of eukaryotic initiation factor 2alpha, Biochemistry 2003; 42:5453-5460.
22. Nanashima N, Asano J, Hayakari M, Nakamura T, Nakano H, Yamada T, Shimizu T, Akita M, Fan Y, Tsuchida S. Nuclear localization of STAT5A modified with O-linked N-acetylglu- cosamine and early involution in the mammary gland of Hirosaki hairless rat. J. Biol. Chem. 2005; 280:43010-43016.
23. Ahmad I, Hoessli DC, Walker-Nasir E, Choudhary MI, Rafik SM, Shakoori AR. Phosphorylation and glycosylation interplay: Protein modifications at hydroxy amino acids and prediction of signaling functions of the human beta(3) integrin family. J. Cell Biochem. 2006; 99:706-718.
24. Kim YH, Song M, Oh YS, Heo K, Choi JW, Park JM, Kim SH, Lim S, Kwon HM, Ryu SH, Suh PG. Inhibition of phospholipase C-beta1-mediated signaling by O-GlcNAc modification. J. Cell. Physiol. 2006; 207:689-696.
25. Musicki B, Kramer MF, Becker RE, Burnett AL. Inactivation of phosphorylated endothelial nitric oxide synthase (Ser-1177) by O-GlcNAc in diabetes-associated erectile dysfunction. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:11870-11875.
26. Holt GD, Hart GW. The subcellular distribution of terminal N-acetylglucosamine moieties. Localization of a novel protein- saccharide linkage, O-linked GlcNAc. J. Biol. Chem. 1986; 261: 8049-8057.
27. Schlummer S, Vetter R, Kuder N, Henkel A, Chen YX, Li YM, Kuhlmann J, Waldmann H. Influence of serine O-glycosylation or O-phosphorylation close to the vJun nuclear localisation sequence on nuclear import. ChemBioChem. 2006; 7:88-97.
28. Zachara NE, Hart GW. The emerging significance of O-GlcNAc in cellular regulation. Chem. Rev. 2002; 102:431-438.
29. Wells L, Whelan SA, Hart GW. O-GlcNAc: A regulatory posttranslational modification. Biochem. Biophys. Res. Commun. 2003; 302:435-441.
30. Wells L, Vosseller K, Hart GW. Glycosylation of nucleocyto-plasmic proteins: Signal transduction and O-GlcNAc. Science 2001; 291:2376-2378.
31. Okuyama R, Marshall S. UDP-N-acetylglucosaminyl transferase (OGT) in brain tissue: temperature sensitivity and subcellular distribution of cytosolic and nuclear enzyme. J. Neurochem. 2003; 86:1271-1280.
32. O’Donnell N, Zachara NE, Hart GW, Marth JD. Ogt-dependent X-chromosome-linked protein glycosylation is a requisite modification in somatic cell function and embryo viability. Mol. Cell. Biol. 2004; 24:1680-1690.
33. Shafi R, Iyer SP, Ellies LG, O’Donnell N, Marek KW, Chui D, Hart GW, Marth JD. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:5735-5739.
34. Lazarus BD, Roos MD, Hanover JA. Mutational analysis of the catalytic domain of O-linked N-acetylglucosaminyl transferase. J. Biol. Chem. 2005; 280:35537-35544.
35. Kreppel LK, Hart GW. Regulation of a cytosolic and nuclear O-GlcNAc transferase. Role of the tetratricopeptide repeats. J. Biol. Chem. 1999; 274:32015-32022.
36. Iyer SP, Akimoto Y, Hart GW. Identification and cloning of a novel family of coiled-coil domain proteins that interact with O-GlcNAc transferase. J. Biol. Chem. 2003; 278:5399-5409.
37. Lazarus BD, Love DC, Hanover JA, Recombinant O-GlcNAc transferase isoforms: identification of O-GlcNAcase, yes tyrosine kinase, and tau as isoform-specific substrates. Glycobiology 2006; 16:415-421.
38. Kreppel LK, Blomberg MA, Hart GW. Dynamic glycosylation of nuclear and cytosolic proteins. Cloning and characterization of a unique O-GlcNAc transferase with multiple tetratricopeptide repeats. J. Biol. Chem. 1997; 272:9308-9315.
39. Whelan SA, Hart GW. Proteomic approaches to analyze the dynamic relationships between nucleocytoplasmic protein glycosylation and phosphorylation. Circ. Res. 2003; 93:1047-1058.
40. Konrad RJ, Zhang F, Hale JE, Knierman MD, Becker GW, Kudlow JE. Alloxan is an inhibitor of the enzyme O-linked N-acetylglucosamine transferase. Biochem. Biophys. Res. Commun. 2002; 293:207-212.
41. Gross BJ, Kraybill BC, Walker S. Discovery of O-GlcNAc transferase inhibitors. J. Am. Chem. Soc. 2005; 127:14588-14589.
42. Dennis RJ, Taylor EJ, Macauley MS, Stubbs KA, Turkenburg JP, Hart SJ, Black GN, Vocadlo DJ, Davies GJ. Structure and mechanism of a bacterial beta-glucosaminidase having O-GlcNAcase activity. Nat. Struct. Mol. Biol. 2006; 13:365-371.
43. Rao FV, Dorfmueller HC, Villa F, Allwood M, Eggleston IM, van Aalten DM. Structural insights into the mechanism and inhibition of eukaryotic O-GlcNAc hydrolysis. Embo. J. 2006; 25:1569-1578.
44. Toleman C, Paterson AJ, Kudlow JE. Location and characterization of the O-GlcNAcase active site. Biochim. Biophys. Acta 2006; 1760:829-839.
45. Cetinbas N, Macauley MS, Stubbs KA, Drapala R, Vocadlo DJ. Identification of Asp174 and Asp175 as the key catalytic residues of human O-GlcNAcase by functional analysis of site-directed mutants. Biochemistry 2006; 45:3835-3844.
46. Toleman C, Paterson AJ, Whisenhunt TR, Kudlow JE. Characterization of the histone acetyltransferase (HAT) domain of a bifunctional protein with activable O-GlcNAcase and HAT activities. J. Biol. Chem. 2004; 279:53665-53673.
47. Comtesse N, Maldener E, Meese E. Identification of a nuclear variant of MGEA5, a cytoplasmic hyaluronidase and a beta-N-acetylglucosaminidase. Biochem. Biophys. Res. Commun. 2001; 283:634-640.
48. Wells L, Gao Y, Mahoney JA, Vosseller K, Chen C, Rosen A, Hart GW. Dynamic O-glycosylation of nuclear and cytosolic proteins: further characterization of the nucleocytoplasmic beta-N-acetylglucosaminidase, O-GlcNAcase. J. Biol. Chem. 2002; 277:1755-1761.
49. Whisenhunt TR, Yang X, Bowe DB, Paterson AJ, Van Tine BA, Kudlow JE. Disrupting the enzyme complex regulating O-GlcNAcylation blocks signaling and development. Glycobiology 2006; 16:551-563.
50. Toleman C, Paterson AJ, Shin R, Kudlow JE. Streptozotocin inhibits O-GlcNAcase via the production of a transition state analog. Biochem. Biophys. Res. Commun. 2006; 340:526-534.
51. Perreira M, Kim EJ, Thomas CJ, Hanover JA. Inhibition of O-GlcNAcase by PUGNAc is dependent upon the oxime stereochemistry. Bioorg. Med. Chem. 2006; 14:837-846.
52. Macauley MS, Whitworth GE, Debowski AW, Chin D, Vocadlo DJ. O-GlcNAcase uses substrate-assisted catalysis: kinetic analysis and development of highly selective mechanism-inspired inhibitors. J. Biol. Chem. 2005; 280:25313-25322.
53. Stubbs KA, Zhang N, Vocadlo DJ. A divergent synthesis of 2-acyl derivatives of PUGNAc yields selective inhibitors of O-GlcNAcase. Org. Biomol. Chem. 2006; 4:839-845.
54. Kim EJ, Perreira M, Thomas CJ, Hanover JA. An O-GlcNAcase-specific inhibitor and substrate engineered by the extension of the N-acetyl moiety. J. Am. Chem. Soc. 2006; 128:4234-4235.
55. Kim EJ, Kang DO, Love DC, Hanover JA. Enzymatic characterization of O-GlcNAcase isoforms using a fluorogenic GlcNAc substrate. Carbohydr. Res. 2006; 341:971-982.
56. Peter-Katalinic J. Methods in enzymology: O-glycosylation of proteins. Methods Enzymol. 2005; 405:139-171.
57. Vosseller K, Trinidad JC, Chalkley RJ, Specht CG, Thalhammer A, Lynn AJ, Snedecor JO, Guan S, Medzihradszky KF, Maltby DA, Schoepfer R, Burlingame AL. O-linked N-acetylglucosamine proteomics of postsynaptic density preparations using lectin weak affinity chromatography and mass spectrometry. Mol. Cell. Proteomics 2006; 5:923-934.
58. Wells L, Vosseller K, Cole RN, Cronshaw JM, Matunis MJ, Hart GW. Mapping sites of O-GlcNAc modification using affinity tags for serine and threonine post-translational modifications. Mol. Cell. Proteomics 2002; 1:791-804.
59. Cieniewski-Bernard C, Bastide B, Lefebvre T, Lemoine J, Mounier Y, Michalski JC. Identification of O-linked N-acetylglucosamine proteins in rat skeletal muscle using two-dimensional gel electrophoresis and mass spectrometry, Mol. Cell. Proteomics 2004; 3:577-585.
60. Tai HC, Khidekel N, Ficarro SB, Peters EC, Hsieh-Wilson LC. Parallel identification of O-GlcNAc-modified proteins from cell lysates. J. Am. Chem. Soc. 2004; 126:10500-10501.
61. Khidekel N, Ficarro SB, Peters EC, Hsieh-Wilson LC. Exploring the O-GlcNAc proteome: direct identification of O-GlcNAc-modified proteins from the brain. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:13132-13137.
62. Vocadlo DJ, Hang HC, Kim EJ, Hanover JA, Bertozzi CR. A chemical approach for identifying O-GlcNAc-modified proteins in cells. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:9116-9121.
63. Nandi A, Sprung R, Barma DK, Zhao Y, Kim SC, Falck JR, Zhao Y. Global identification of O-GlcNAc-modified proteins. Anal. Chem. 2006; 78:452-458.
See Also
Chemical Tools for Studying Glycans
Chemistry of Glycans
Cytosolic Glycosylation in Signaling
Identification of Post-Translational Modifications,
Regulating Protein Function by Post-Translational Modification