MCAT Biology and Biochemistry: New for MCAT 2015 (2014)
Chapter 5. Molecular Biology
It was once thought that simple living organisms were generated spontaneously from nonliving matter. When a steak went bad and became infested with larvae, it was because the decomposing meat actually became squirming worms. Most religions have traditional explanations for the origin of human life, too. Children are derived from adults due to the will of a deity; the original adults were placed on the earth by that deity. But as empiricism developed during the Enlightenment, rigorous experiments were used to explain life, resulting in “scientific” models that are gradually replacing more traditional explanations.
One early conclusion was that simple organisms were derived not from decomposing matter but from parental organisms. Subsequently, it was found that some organisms are too small to be seen with the naked eye. These “germs” were eventually implicated as the cause of most major diseases. Gradually the scientific community came to the conclusion that all life was derived from other life. The patterns of inheritance and evolution were elucidated by a chain of scientists, from Mendel through Darwin. But the mechanism remained a mystery. Finally, cellular biology advanced to the point that scientists were aware of two substances found in cells which seemed appropriate vehicles for the transmission of inherited information: DNA and protein. The extreme length and orderly arrangement of repeating units in DNA and protein made it seem very likely that they could contain information. Researchers had waded through a chemical ocean of alphabet soup and suddenly come upon long strings of what looked like letters.
This is where biology stood in the early 1940s. In the ’40s and ’50s, two monumental achievements in microbiology finally clarified the gears in the clock of evolution and how they turn. One was the elucidation of the structure of DNA by Watson and Crick. The other was the proof by Avery, Herriott, Hershey, Chase, and their coworkers that DNA was the fundamental unit of genetic inheritance in microorganisms. In the following discussion, we will summarize the wealth of information that has been built upon these two prescient cornerstones.
5.1 DNA STRUCTURE
General Overview
Understanding the structure of DNA provides great insight into its function, so let’s start at the smallest level and work our way up. DNA is short for deoxyribonucleic acid. DNA and RNA (ribonucleic acid) are called nucleic acidsbecause they are found in the nucleus and possess many acidic phosphate groups.
The building block of DNA is the deoxyribonucleoside 5 triphosphate (dNTP, where N represents one of the four basic nucleosides). Deoxyadenosine 5 triphosphate (dATP) is shown in Figure 1. Deoxyribonucleotides are built from three components. The first is a simple monosaccharide, ribose. [What modification makes this ribose special?1] In a dNTP, carbons on the ribose are referred to as 1, 2, etc. The next component of the dNTP is an aromatic, nitrogenous base, namely adenine (A), guanine (G), cytosine (C), or thymine (T); see Figure 2. (Don’t mix up the DNA base thymine with vitamin B1, thiamine.) These aromatic molecules are bases because they contain several nitrogens which have free electron pairs capable of accepting protons. G and A are derived from a precursor called purine, so they are referred to as the purines. C and T are the pyrimidines.2
A nucleoside is ribose with a purine or pyrimidine linked to the 1 carbon in a β-N-glycosidic linkage. [In the β-N-glycosidic linkage of a nucleoside, is the aromatic base above or is it below the plane of ribose in a Haworth projection?3] The nucleosides are named as follows: A-ribose = adenosine, G-ribose = guanosine, C-ribose = cytidine, T-ribose = thymidine, and U-ribose = uridine. Both purines and pyrimidines have abundant hydrogen bonding potential. [Will adenine and thymine H-bond with each other in dilute aqueous solution (0.1 M, for example)?4]
The final component of the deoxyribonucleotide building block of DNA is a phosphate group. Nucleotides are phosphate esters of nucleosides, with one, two, or three phosphate groups joined to the ribose ring by the 5 hydroxy group. When nucleotides contain three phosphate residues, they may also be referred to as deoxynucleoside triphosphates; they are abbreviated dNTP, where d is for deoxy and N is for nucleoside. In individual nucleotides, N is replaced by A, G, C, T, or U. Because they contain acidic phosphates, the nucleotides may also be referred to by a name ending in “ylate.” For example, TTP is thymidylate. The ubiquitous energy molecule, ATP, is a nucleotide which may be called adenylate (it’s not deoxy).
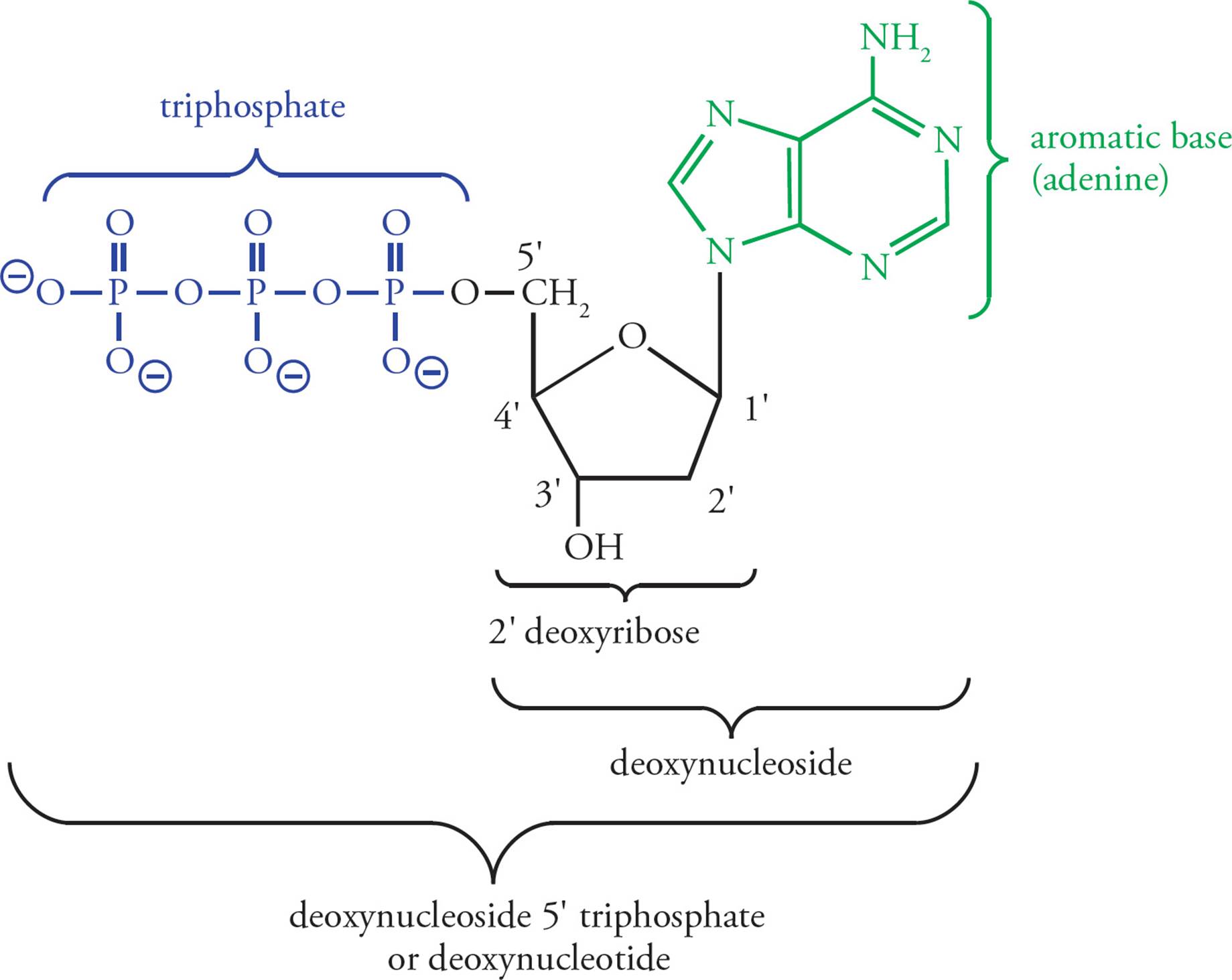
Figure 1 Deoxyadenosine Triphosphate (dATP)
The ribose + phosphate portion of the nucleotide is referred to as the backbone of DNA, because it is invariant. The base is the variable portion of the building block. Hence there are four different dNTPs, and they differ only in the aromatic base. [What is the backbone in protein, and what is the variable portion of the amino acid?5 If an enzyme binds to a specific sequence of nucleotides in DNA, will the binding specificity be derived from interactions of portions of the polypeptide enzyme with the ribose and phosphate groups or with the purine and pyrimidine bases?6]
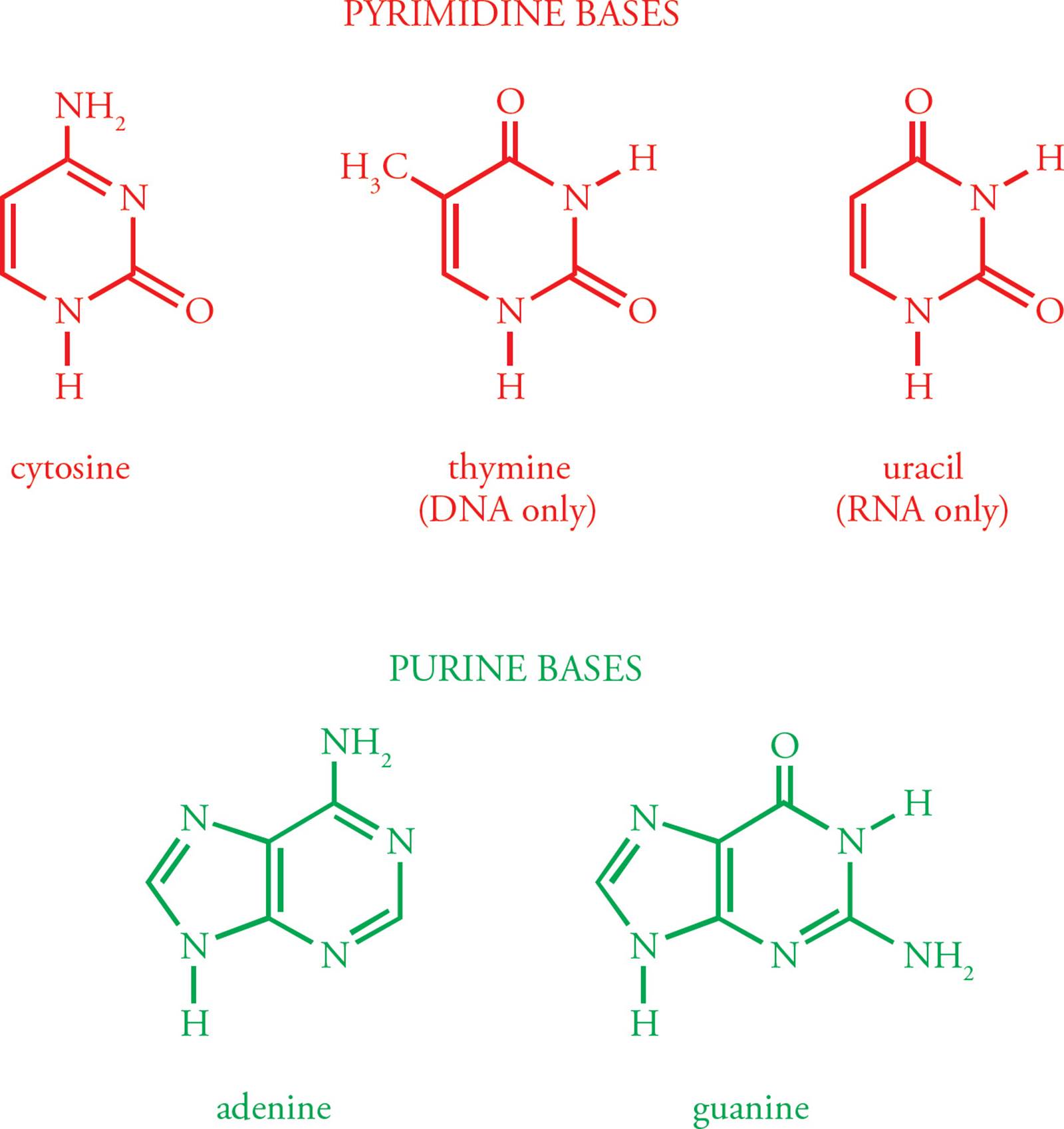
Figure 2 Aromatic Bases of DNA and RNA
Polynucleotides
Nucleotides in the DNA chain are covalently linked by phosphodiester bonds between the 3 hydroxy group of one deoxyribose and the 5 phosphate group of the next deoxyribose (Figure 3). [Which reaction is more thermodynamically favorable: the polymerization of nucleoside monophosphates, or the polymerization of nucleoside triphosphates?7] A polymer of several nucleotides linked together is termed an oligonucleotide, and a polymer of many nucleotides is a polynucleotide. Since the only unique part of the nucleotide is the base, the sequence of a polynucleotide can be abbreviated by simply listing the bases attached to each nucleotide in the chain. The end of the chain with a free 5 phosphate group is written first in a polynucleotide, with other nucleotides in the chain indicated in the 5 to 3 direction. [Which of the nucleotides in the oligonucleotide ACGT has a free 3 hydroxy group?8]
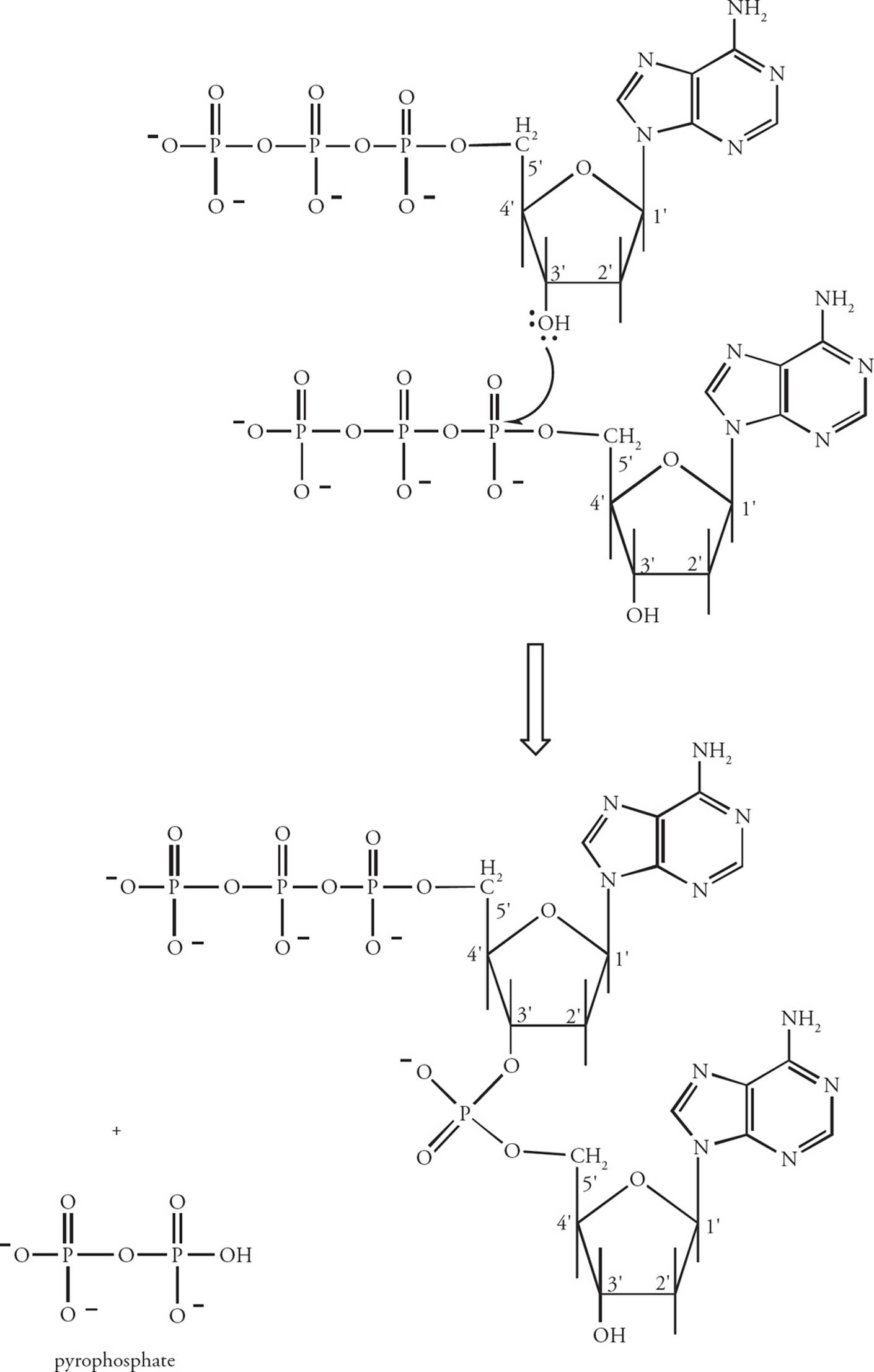
Figure 3 The Polymerization of Nucleotides
The Watson-Crick Model of DNA Structure
James Watson and Francis Crick (with the help of Maurice Wilkins and Rosalind Franklin) developed a model of the structure of DNA in the cell. According to the Watson-Crick model, cellular DNA is a right-handed double helix held together by hydrogen bonds between bases. It is important to understand each facet of this model.
In the cell, DNA does not exist in the form of a single long polynucleotide. Instead, the DNA found in the nucleus is double-stranded (ds). In ds-DNA, two very long polynucleotide chains are hydrogen-bonded together in an antiparallel orientation. Antiparallel means the 5 end of one chain is paired with the 3 end of the other. [What common protein structure often depends on H-bonds between antiparallel chains?9] The H-bonds in ds-DNA are between the bases on adjacent chains. This H-bonding is very specific: A is always H-bonded to T, and G is always H-bonded to C (Figure 4). Note that this means an H-bonded pair always consists of a purine plus a pyrimidine.10Thus both types of base pairs (AT or GC) take up the same amount of room in the DNA double helix. The GC pair is held together by three hydrogen bonds, the AT pair by two. Two chains of DNA are said to be complementary if the bases in each strand can hydrogen bond when the strands are oriented in an antiparallel fashion. If we are talking about ds-DNA 100 nucleotides long, we would say it is 100 base pairs (bp) long. A kbp (kilobase pair) is ds-DNA 1000 nucleotides long.
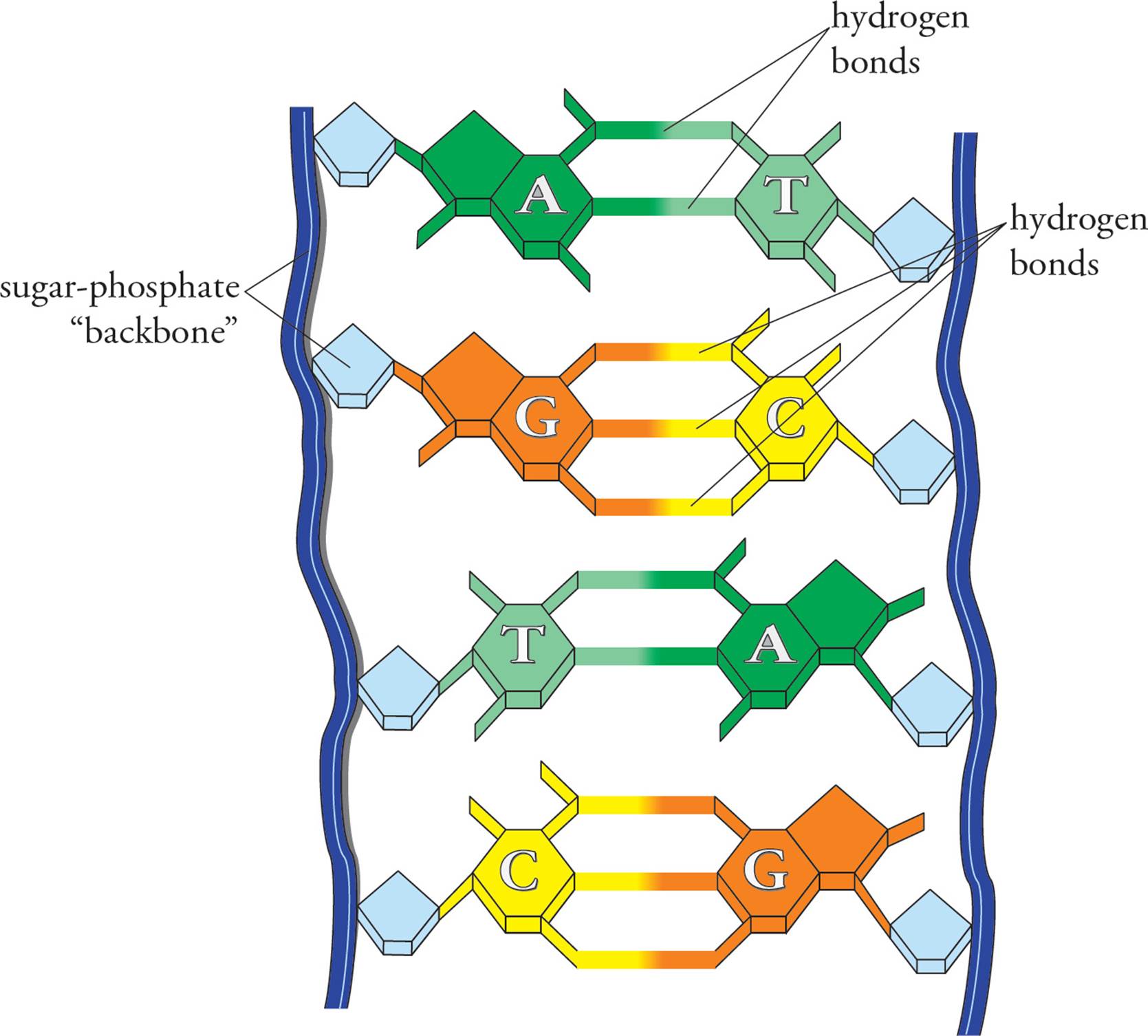
Figure 4 Base Pairing
The binding of two complementary strands of DNA into a double-stranded structure is termed annealing, or hybridization. The separation of strands is termed melting, or denaturation. The temperature at which a solution of DNA molecules is 50 percent melted is termed the Tm. [Would the Tm of ATTATCAT and its complementary strand be higher than, lower than, or equal to the melting temperature of AGTCGCAT and its complementary strand?11 If you attached methyl groups to all the acidic phosphate oxygens along the length of a DNA double helix, would the chain have a higher or lower Tm than normal DNA?12]
• Which of the following is/are true about ds-DNA?
I. If the amount of G in a double helix is known, the amount of C can be calculated.
II. If the fraction of purine nucleotides and the total molecular weight of a double helix are known, the amount of cytosine can be calculated.
III. The two chains in a piece of ds-DNA containing mostly purines will be bonded together more tightly than the two chains in a piece of ds-DNA containing mostly pyrimidines.
IV. The oligonucleotide ATGTAT is complementary to the oligonucleotide ATACAT.13
There is another important detail about DNA structure: Not only is it double stranded, it is also coiled. In ds-DNA, the two hydrogen-bonded antiparallel DNA strands form a right-handed double helix (meaning it corkscrews in a clockwise motion) with the bases on the interior and the ribose/phosphate backbone on the exterior. The double helix is stabilized by van der Waals interactions between the bases, which are stacked upon each other. Hydrophobic interactions between the bases are also very important in stabilizing the double helix. [But wait a minute. “Hydrophobic interactions between bases?” Isn’t that a contradiction in terms? How can a base be hydrophobic?14] The bases lie in a plane, perpendicular to the length of the DNA molecule, stacked 3.4 angstroms (Å) apart from each other. The helix pattern repeats itself (i.e., completes a full turn) once every 34 angstroms, which is every 10 base pairs. While the length of a DNA double helix may vary enormously, from a few Å in an oligonucleotide to macroscopic lengths in a chromosome, the width is always 20 Å. [If a human chromosome has 9 × 107 base pairs, how long would the chromosome be if it were stretched out completely?15]
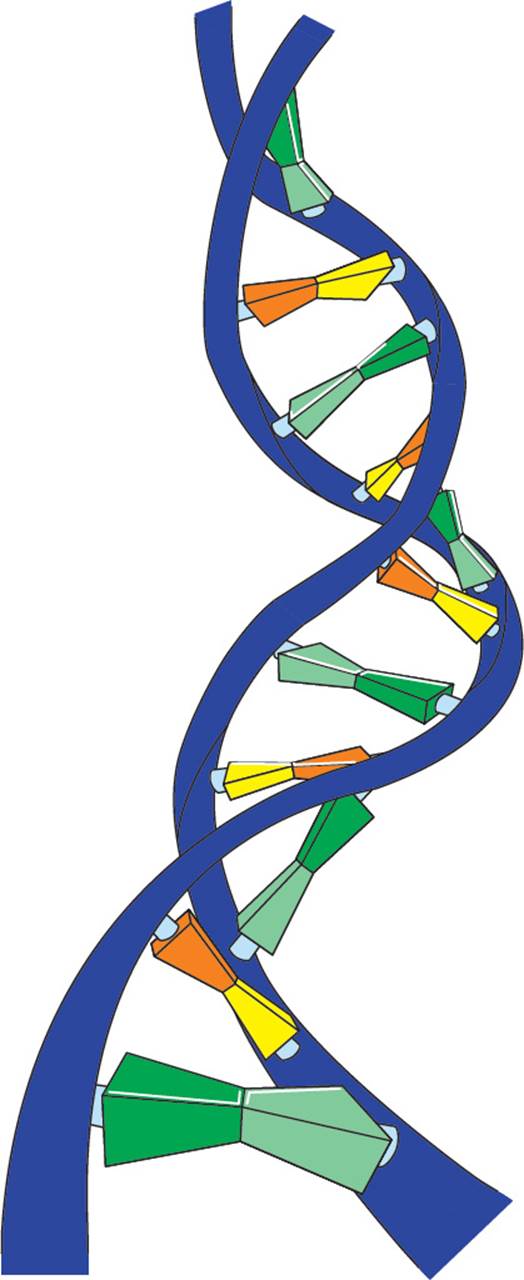
Figure 5 A Small Section of a DNA Double Helix
Chromosome Structure and Packing
The sum total of an organism’s genetic information is called its genome. Eukaryotic genomes are composed of several large pieces of linear ds-DNA; each piece of ds-DNA is called a chromosome. Humans have 46 chromosomes, 23 of which are inherited from each parent. Prokaryotic (bacterial) genomes are composed of a single circular chromosome. Viral genomes may be linear or circular DNA or RNA. The human genome consists of over 109 base pairs while bacterial genomes contain only 106 base pairs. But there is no direct correlation between genome size and evolutionary sophistication, since the organisms with the largest known genomes are amphibians. Much of the size difference in higher eukaryotic genomes is the result of repetitive DNA that has no known function.
If the DNA remained as a simple double helix floating free in the cell, it would be very bulky and fragile. Prokaryotes have a distinctive mechanism for making their single circular chromosome more compact and sturdy. An enzyme called DNA gyrase uses the energy of ATP to twist the gigantic circular molecule. Gyrase functions by breaking the DNA and twisting the two sides of the circle around each other. The resulting structure is a twisted circle that is composed of ds-DNA. As discussed above, the two strands are already coiled, forming a helix. The twists created by DNA gyrase are called supercoils, since they are coils of a structure that is already coiled.
Since eukaryotes have even more DNA in their genome than prokaryotes, the eukaryotic genome requires denser packaging to fit within the cell (Figure 6). To accomplish this, eukaryotic DNA is wrapped around globular proteins called histones. After being wrapped around histones, but before being completely packed away, DNA has the microscopic appearance of beads on a string. The beads are called nucleosomes; they are composed of DNA wrapped around an octamer of histones (a group of eight). The octamer is composed of two units of each of the histone proteins H2A, H2B, H3 and H4. The string between the beads is a length of double-helical DNA called linker DNA and is bound by a single linker histone. Fully packed DNA is called chromatin; it is composed of closely stacked nucleosomes. [Based on your knowledge of the interactions of macromolecules and the chemical composition of DNA, do you suppose that histones mostly basic or mostly acidic?16]
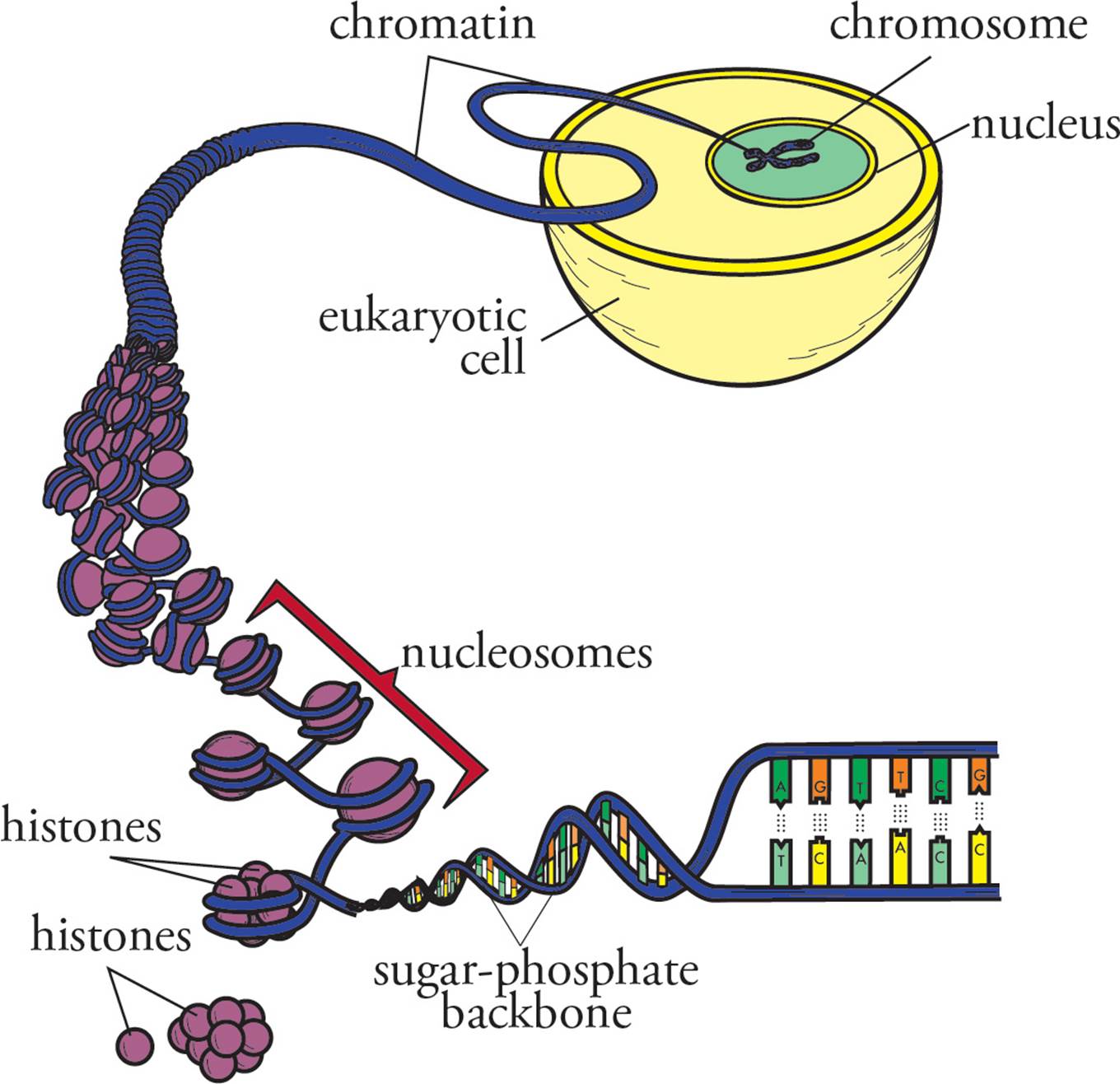
Figure 6 DNA Packaging
The following flow summarizes the structure of DNA in the nucleus: Deoxyribose → add base → nucleoside → add three phosphates → nucleotide → polymerize with loss of two phosphates → oligonucleotide → continue polymerization → single stranded polynucleotide → two complete chains H-bond in antiparallel orientation → ds DNA chain → coiling occurs → ds helix → wrap around histones → nucleosomes → complete packaging → chromatin. Remember, each individual double-stranded piece of chromatin is condensed into a chromosome during mitosis and meiosis (see Chapters 7 and 8).
To look for patterns and morphology, chromosomes can be stained with chemicals. Usually, condensed metaphase chromosomes are used, as they are compact and easier to see. When chromosomes are treated, distinct light and dark regions become visible. The darker regions are denser, and are called heterochromatin. Heterochromatin is rich in repeats (see below). The lighter regions are less dense and are called euchromatin. Density gives a sense of DNA coiling or compactness, and these patterns are constant and heritable. It’s now known that the lighter regions have higher transcription rates and therefore higher gene activity. The looser packing makes DNA accessible to enzymes and proteins.
Giemsa stain can also be used, and produces what are called “G-banding patterns”. Here too, darker staining regions are more dense than lighter staining regions. Chromosome bands are constant and specific to each chromosome, which means they can be used for diagnostic purposes (where cytologists look at chromosome structure). Banding patterns have also been linked to DNA replication, as it’s been shown that lighter staining regions start replication earlier than darker staining regions. Again, this is likely due to accessibility of the DNA.
Centromeres
A centromere is the region of the chromosome to which spindle fibers attach during cell division. The fibers attach via kinetochores, multiprotein complexes that act as anchor attachment sites for spindle fibers. Other protein complexes also bind the centromere after DNA replication to keep sister chromatids attached to each other. Centromeres are made of heterochromatin, and repetitive DNA sequences. Chromosomes have p (short) and q (long) arms, and centromere position defines the ratio between the two (Figure 7).
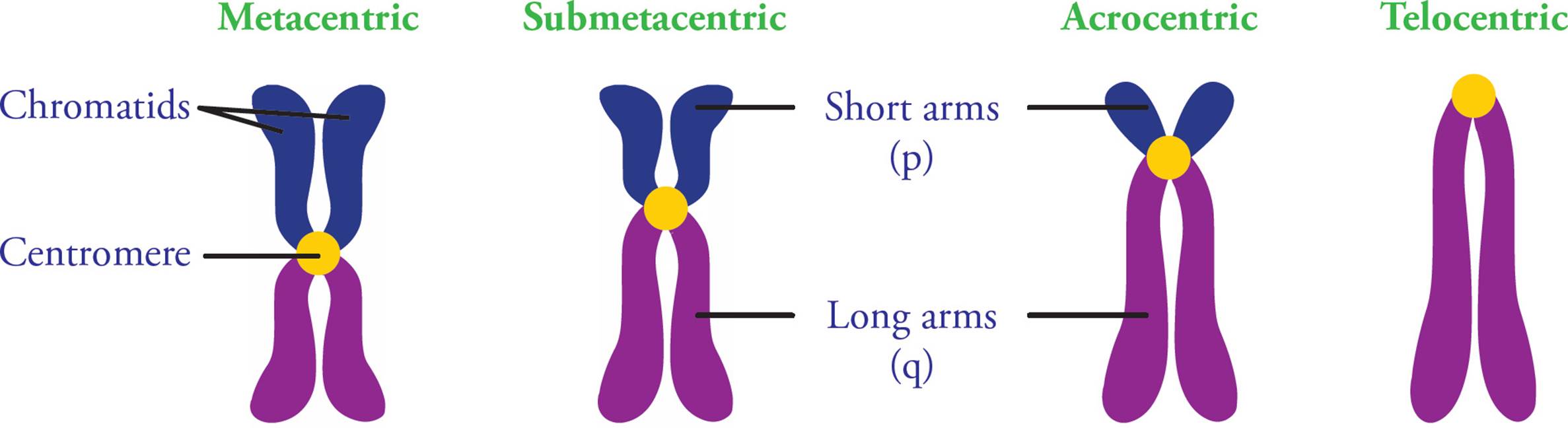
Figure 7 Centromere Positions
Telomeres
The ends of linear chromosomes are called telomeres. At the DNA level, these regions are distinguished by the presence of distinct nucleotide sequences repeated 50 to several hundred times. The repeated unit is usually 6-8 base pairs long and guanine-rich. Many vertebrates (including humans and mice) have the same repeat: 5′-TTAGGG-3′. Telomeres are composed of both single and double stranded DNA. Single stranded DNA is found at the very end of the chromosome and is about 300 base pairs in length. It loops around to form a knot, held together by many telomere-associated proteins. This stabilizes the end of the chromosome; specialized telomere cap proteins distinguish telomeres from double stranded breaks (Section 5.4), and this prevents activation of repair pathways.
Telomeres function to prevent chromosome deterioration and also prevent fusion with neighboring chromosomes. They function as disposable buffers, blocking the ends of chromosomes. DNA replication of telomeres represents a special challenge to cellular machinery (see Section 5.4). Since most prokaryotes have circular genomes, their DNA does not contain telomeres.
5.2 GENOME STRUCTURE AND GENOMIC VARIATIONS
The human genome contains 24 different chromosomes (22 autosomes, plus two different sex chromosomes), 3.2 billion base pairs, and codes for about 21,000 genes. The sequence of the human genome was reported by two independent groups in 2001 (the publicly funded Human Genome Project lead by Dr. Francis Collins, and Dr. J. Craig Venter and his firm Celera Genomics).
The human genome has numerous regions with high transcription rates, separated by long stretches of intergenic space. Intergenic regions are composed of noncoding DNA; they may direct the assembly of specific chromatin structures, and can contribute to the regulation of nearby genes, but many have no known function. Tandem repeats and transposons (see below) are major components of intergenic regions.
Genomic regions with high transcription rates are rich in genes. A gene is a DNA sequence that encodes a gene product. It includes both regulatory regions (such as promoters and transcription stop sites), and a region that codes for either a protein or a non-coding RNA (see Section 5.7).
Nucleotide Variation
Small scale and large scale variation across a genome is common. For example, one person could have the sequence CCCGGG, while another has CCTGGG. It’s been predicted that there are single nucleotide changes once in every 1,000 base pairs in the human genome. These variations are called single nucleotide polymorphisms (SNPs, pronounced “snips”) and are essentially mutations. [If the size of the human genome is just over 3 billion base pairs, approximately how many human SNPs are there?17] These SNPs occur most frequently in noncoding regions of the genome, however some SNPs can lead to specific traits and phenotypes. For example, about 70% of people taste phenylthiocarbamide (PTC) as very bitter, and the remaining 30% don’t taste PTC at all. You may have done this test yourself, since PTC response is commonly used as an example in genetics classes. This ability is a dominant genetic trait and is determined by a gene on chromosome 7. Three SNPs in this gene determine PTC taste sensitivity.
Copy Number Variation
Copy-number variations (CNVs) are structural variations in the genome that lead to different copies of DNA sections. Large regions of the genome (103 to 106 base pairs) can be duplicated (increasing copy number) or deleted (decreasing copy number). The specific mechanism by which this occurs is not clear, but it may be due to misalignment of repetitive DNA sequences during synapsis of homologous chromosomes in meiosis. These changes therefore apply to much larger regions of the genome compared to SNPs. They are a normal part of our genome (0.4% of the genome can have CNV), but have also been associated with cancer and other diseases. Genes involved in immune system function, as well as brain development and activity, are often enriched in CNVs.
Repeated Sequences: Tandem Repeats
Much of our genome is single copy, meaning there is one copy of the gene in a haploid set of the genome. This is true for most eukaryotic genes that code for proteins. However, genomes also have regions of tandem repeats, where short sequences of nucleotides are repeated one right after the other, from as little as three to over 100 times. The human genome has over a thousand regions of tandem repeats. Repeats can be unstable, when the repeating unit is short (such as di- or trinucleotides) or when the repeat itself is very long. Unstable tandem repeats can lead to chromosome breaks and some have been implicated in disease. Tandem repeats often show variations in length between individuals, which can be useful in DNA fingerprinting (see Appendix I). Heterochromatin, centromeres and telomeres are all rich in repeats.
Repeated Sequences: Transposons
Both prokaryotes and eukaryotes have mobile genetic elements in their genomes, called transposable elements or transposons. It is thought that many eukaryotic transposons are degenerate (old and defective) retroviruses. “Genetic mobility” means that these short segments can jump around the genome. Transposons can cause mutations and chromosome changes (such as inversions, deletions and rearrangements) and these will be discussed in Section 5.5.
There are three common types of transposons, each with a different structure. The first type is the simplest and is called an IS element (Figure 8, top). It is composed of a transposase gene (discussed below), flanked by inverted repeat sequences. The structure of an example inverted repeat is shown in Figure 9. Some transposons are more complex, in that they also contain additional genes (Figure 8, middle). For example, some transposons contain genes for antibiotic resistance. Finally, composite transposons have two similar or identical IS elements with a central region in between (Figure 8, bottom).
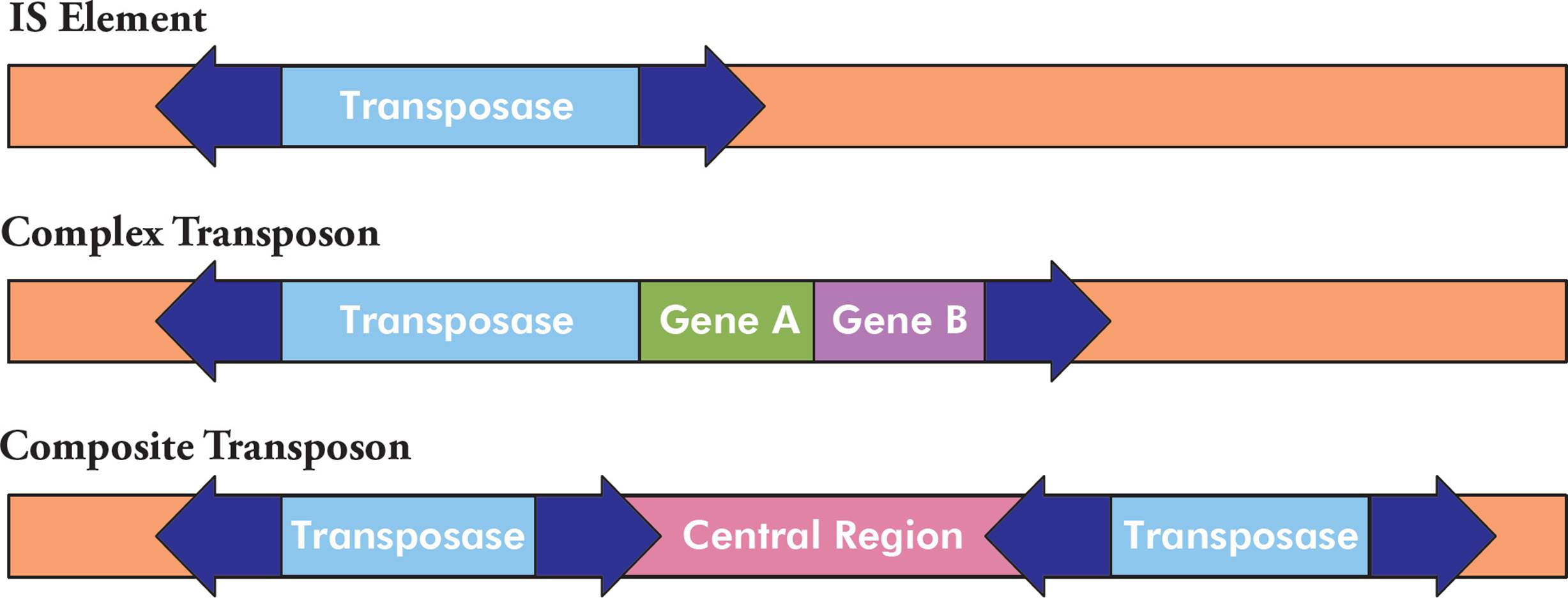
Figure 8 Transposon Structure
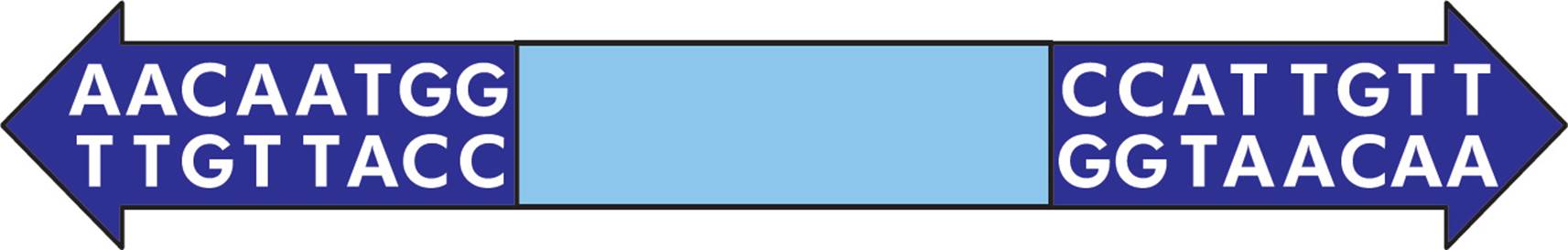
Figure 9 Inverted Repeats
All transposons contain a gene that codes for a protein called transposase. This enzyme has “cut and paste” activity, where it catalyzes mobilization of the transposon (excision from the donor site) and integration into a new genetic location (the acceptor site). Sometimes the transposon sequence is completely excised and moved, and sometimes it is duplicated and moved, while still maintained at the original location (Figure 10). The inverted repeats are important for this mobilization.
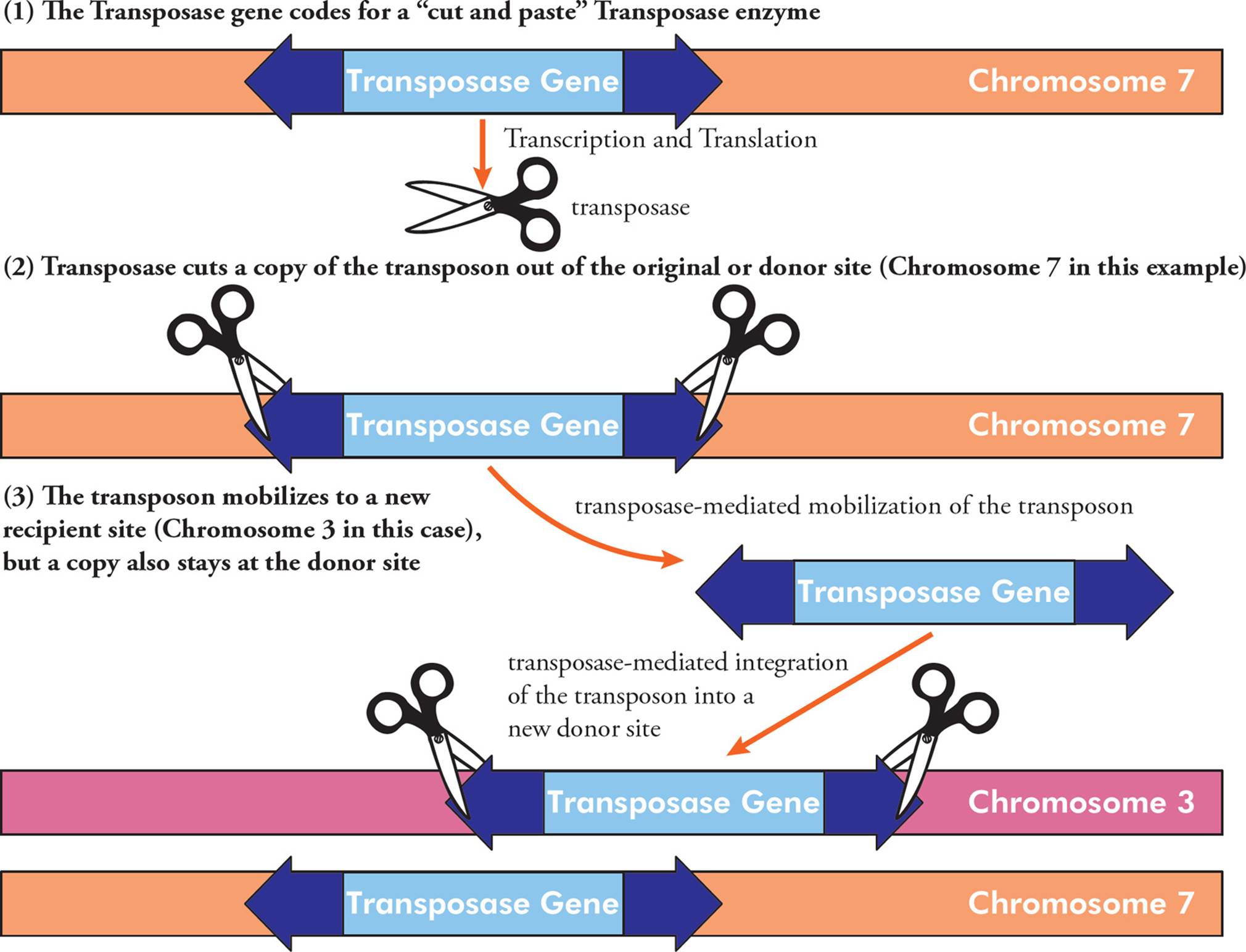
Figure 10 The Mechanism of Transposon Mobilization
Many mobilizations have no effect because the transposon inserts into a relatively unimportant part of the genome. However, transposons can cause mutations if they jump into an important part of the genome (Section 5.5).
5.3 THE ROLE OF DNA
DNA encodes and transmits the genetic information passed down from parents to offspring. Before 1944 it was generally believed that protein, rather than DNA, carried genetic information, since proteins have an “alphabet” of 20 letters (the amino acids), while DNA’s “alphabet” has only 4 letters (the four nucleotides). But in that year, Oswald Avery showed that DNA was the active agent in bacterial transformation. In short, this means he proved that pure DNA from one type of E. coli bacteria could transform E. coli of another type, causing it to acquire the genetic nature of the first type. Later Hershey and Chase proved that DNA was the active chemical in the infection of E. colibacteria by bacteriophage T2.18 These experiments will be discussed in more detail in Chapter 8.
The Genetic Code
DNA does not directly exert its influence on cells, but merely contains sequences of nucleotides known as genes that serve as templates for the production of another nucleic acid known as RNA. The process of reading DNA and writing the information as RNA is termed transcription. This can generate either a final gene product (as in the case of all non-coding RNAs, discussed below), or a messenger molecule. The messenger RNA (mRNA) is then read, and the information is used to construct protein. The synthesis of proteins using RNA as a template is termed translation, and is accomplished by the ribosome, which is a massive enzyme composed of many proteins and pieces of RNA (known as ribosomal RNA or rRNA).19
The overall process looks like this: DNA → RNA → protein. This unidirectional flow equation represents the Central Dogma (fundamental law) of molecular biology. This is the mechanism whereby inherited information is used to create actual objects, namely enzymes and structural proteins.
This language used by DNA and mRNA to specify the building blocks of proteins is known as the Genetic Code. The alphabet of the genetic code contains only four letters (A, T, G, C). How can four letters specify the ingredients of the multitude of proteins in every cell? [What is the smallest “word” size that would allow this four-letter alphabet to encode twenty different amino acids?20] A number of experiments confirmed that the genetic code is written in three-letter words, each of which codes for a particular amino acid. A nucleic acid word (3 nucleotide letters) is referred to as a codon.
The genetic code is represented in Figure 11. The first nucleotide in a codon is given at the left, the second on top, and the third on the right. At the intersection of these three nucleotides is the amino acid called for by that codon. [Why is uracil (U) shown in the chart, and why is thymine (T) absent?21 The codon GTG in DNA is transcribed in RNA as __, which the ribosome translates into what amino acid?22]
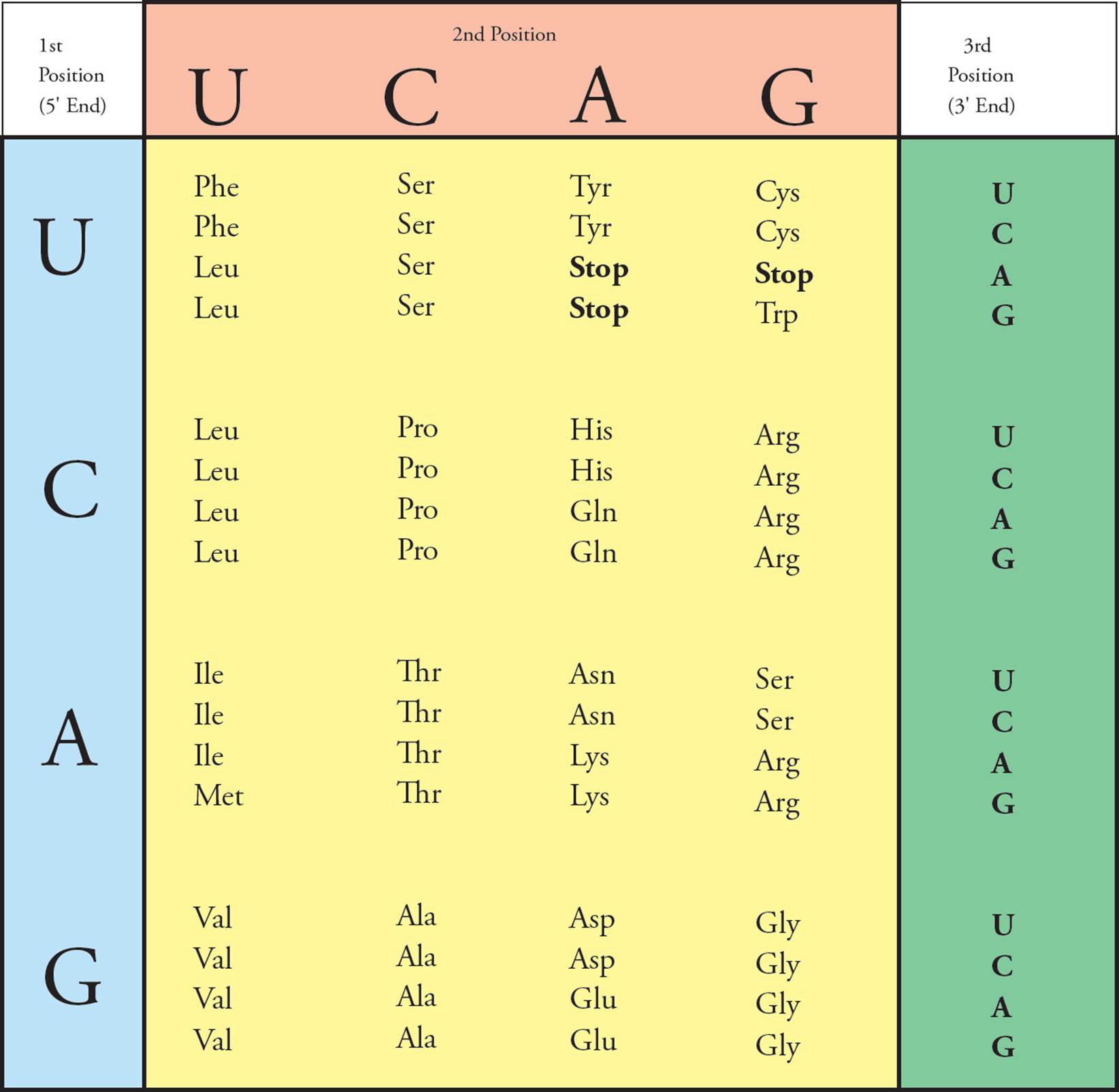
Figure 11 The Genetic Code
• The genetic code was studied by experimenters using a cell-free protein synthesis system. All of the materials necessary for protein synthesis (ribosomes, amino acids, tRNA, GTP, ATP) were purified and placed in a beaker. Then synthetic RNA was added, and protein was translated from this template. For example, when synthetic RNA containing only cytosine (CCCCC …) was added, polypeptides containing only proline (polyproline) resulted. What kind of synthetic RNA would give rise to a mixture of polyproline, polyhistidine, and polythreonine?23
There are 64 codons. Sixty-one of them specify amino acids; the remaining three are called stop codons. Their function is to notify the ribosome that the protein is complete and cause it to stop reading the mRNA (see Section 5.5). Stop codons are also called nonsense codons, since they don’t code for any amino acid. Note that most of the twenty amino acids can be coded for by more than one codon. Often, all four of the codons with the same first two nucleotides (e.g., CU_) encode the same amino acid. [If the last nucleotide in the codon CUU is changed in a gene that codes for a protein, will the protein be affected?24] Two or more codons coding for the same amino acid are known as synonyms. Because it has such synonyms, the genetic code is said to be degenerate. However, it is very important to realize that though an amino acid may be specified by several codons, each codon specifies only a single amino acid. This means that each piece of DNA can be interpreted only one way: The code has no ambiguity.
The code in Figure 11 is the standard genetic code and is used by most organisms. However, some protists use an alternate genetic code, and the mitochondrial genome (see Section 5.10) of many organisms (including humans and many other vertebrates) uses a slightly different code.
Beyond the Central Dogma
There are several aspects of molecular biology that aren’t explicitly stated in the Central Dogma.
• Some viruses (retroviruses) make DNA from RNA using the enzyme reverse transcriptase (see Chapter 6).
• Information can also be transferred in other ways. For example, DNA methylation and post-translational modification of proteins can alter gene expression and convey information, despite the fact that neither is directly included in the Central Dogma.
• Many final gene products are not proteins, but are RNAs instead.
5.4 DNA REPLICATION
The DNA genome is the control center of the cell. When mitosis produces two identical daughter cells from one parental cell, each daughter must have the same genome as the parent. Hence, cell division requires duplication of the DNA, known as replication. This is an enzymatic process, just as the Krebs cycle and glycolysis are enzymatic processes. It occurs during S (synthesis) phase in interphase of the cell cycle (Chapter 7). Let’s go through the process of replication, stopping to add essential facts to a list of things to memorize. But before we get bogged down with details, we should have a look at the big picture.
There is only one logical way to make a new piece of DNA that is identical to the old one: copy it. The old DNA is called parental DNA, and the new is called daughter DNA. What is the relationship between parental and daughter DNA after replication? There are several possibilities (Figure 12). In other words, where do the atoms from the parent go when the daughters are made?
Experiments done by Meselson and Stahl in 1958 aimed to determine if DNA replication is semiconservative, conservative, or dispersive (Figure 12). In conservative replication, the parental ds-DNA would remain as-is while an entirely new double-stranded genome was created. The dispersive theory said that both copies of the genomes were composed of scattered pieces of new and old DNA. Meselson and Stahl showed that replication is semiconservative; after replication, one strand of the new double helix is parental (old) and one strand is newly synthesized daughter DNA.
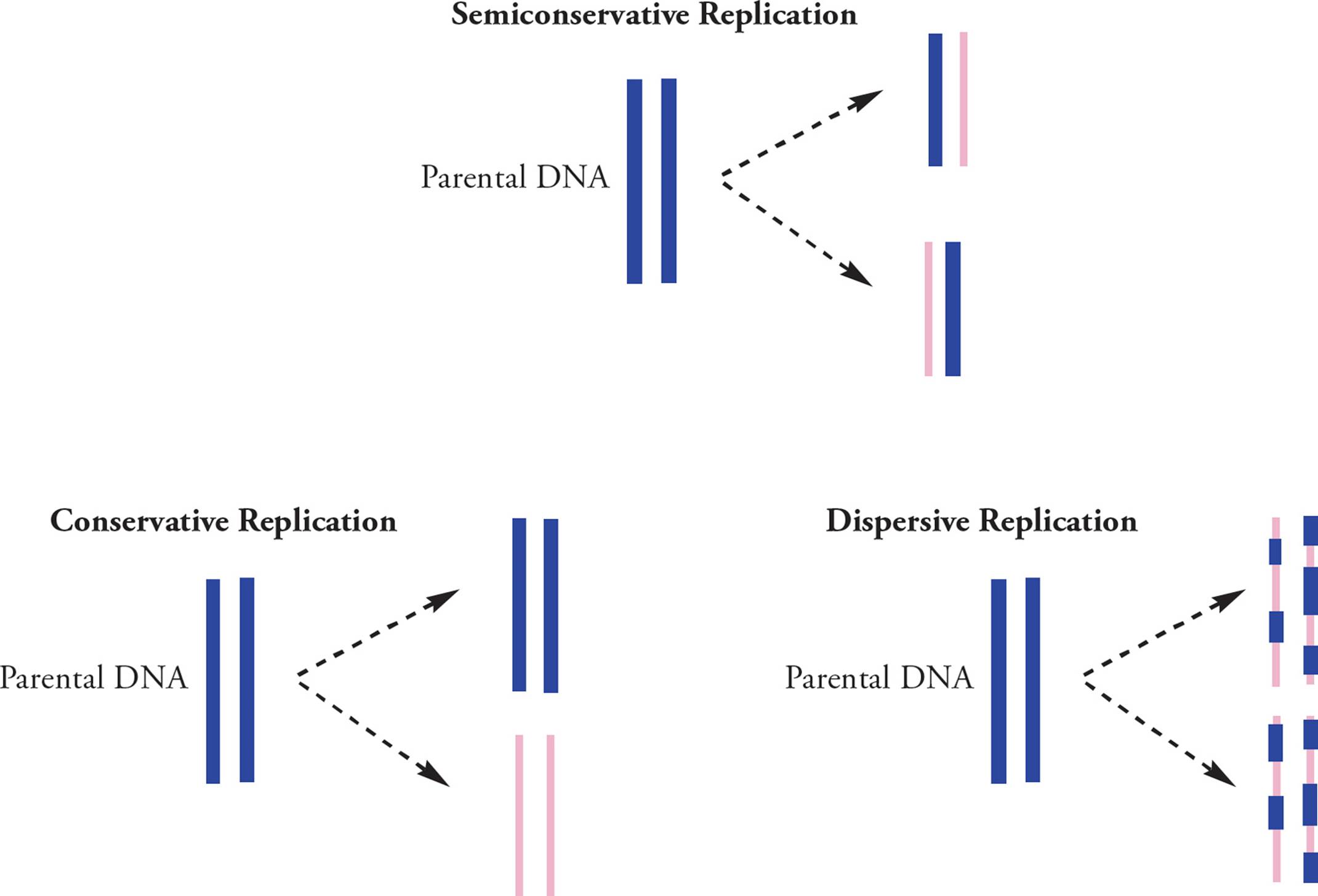
Figure 12 Meselson-Stahl Experiments
Let’s begin the list of things to memorize here:
1) DNA replication is semiconservative.
Individual strands of the double-stranded parent are pulled apart, then a new daughter strand is synthesized using the parental DNA as a template to copy from.25 Each new daughter chain is perfectly __26 to its template or parent.
Now we’ll look at replication at the molecular level. When it is not being replicated, DNA is tightly coiled. The replication process cannot begin unless the double helix is uncoiled and separated into two single strands. The enzyme that unwinds the double helix and separates the strands is called helicase. [Would you expect helicase to use the energy of ATP hydrolysis to do its job?27] The place where the helicase begins to unwind is not random. It is a specific location (sequence of nucleotides) on the chromosome called the origin of replication (abbreviated ORI). This sequence is found by proteins with tertiary structures to specifically recognize a particular pattern of nucleotides. They scan along the chromosome (like a train on a track) until they find the right spot, then they call in helicase and other enzymes to initiate DNA replication. In prokaryotes, a protein called DnaA finds the ORI to initiate DNA replication. In eukaryotes, three proteins cooperate to find the ORI, two of which are synthesized during M and G1 phases of the cell cycle (see Chapter 7), but are rapidly destroyed once the S phase begins. This means these two proteins link DNA replication to the cell cycle, ensuring DNA replication doesn’t initiate during other phases of the cell cycle.
When helicase unwinds the helix at the origin of replication, the helix gets wound more tightly upstream and downstream from this point.28 The chromosome would get tangled and eventually break, except that enzymes called topoisomerases cut one or both of the strands and unwrap the helix, releasing the excess tension created by the helicases. Another potential problem is that single-stranded DNA is much less stable than ds-DNA. Single-strand binding proteins (SSBPs) protect DNA that has been unpackaged in preparation for replication and help keep the strands separated. The separated strands are referred to as an open complex. Replication may now begin.
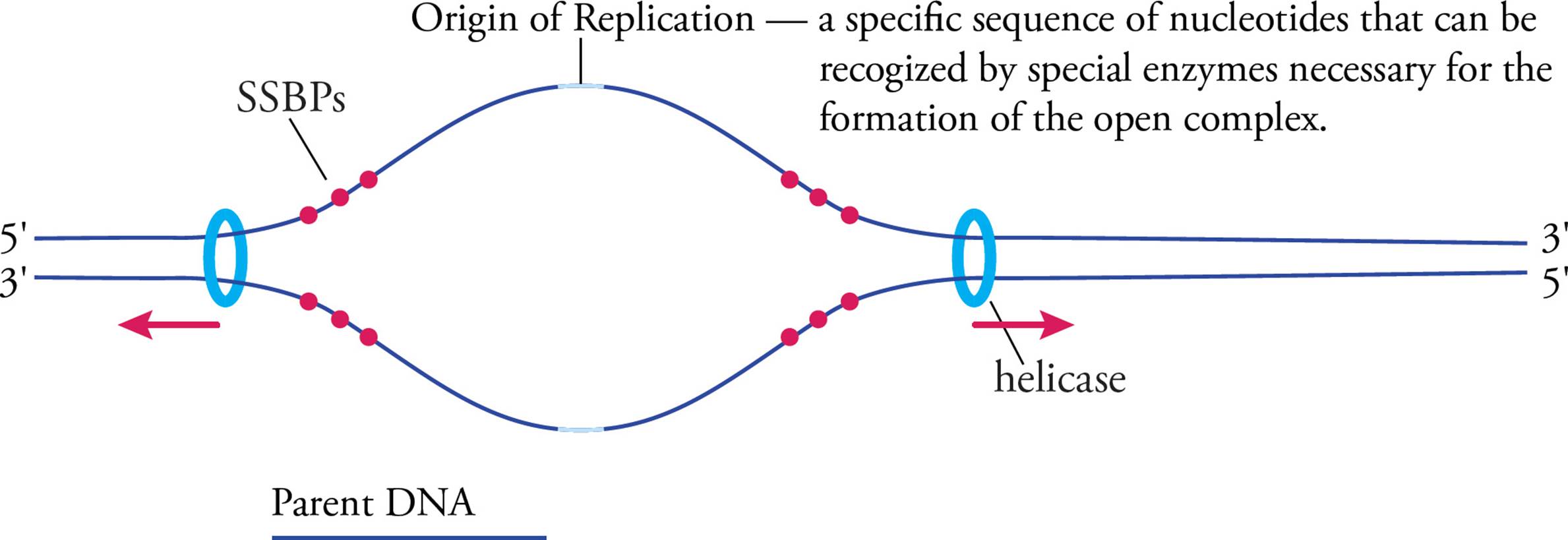
Figure 13 Initiation—The Open Complex
An RNA primer must be synthesized for each template strand. This is accomplished by a set of proteins called the primosome, of which the central component is an RNA polymerase called primase. Primer synthesis is important because the next enzyme, DNA polymerase, cannot start a new DNA chain from scratch. It can only add nucleotides to an existing nucleotide chain. The RNA primer is usually 8–12 nucleotides long, and is later replaced by DNA.
Daughter DNA is created as a growing polymer. DNA polymerase (DNA pol) catalyzes the elongation of the daughter strand using the parental template, and elongates the primer by adding dNTPs to its 3′ end. In fact, the 3′ hydroxyl group acts as a nucleophile in the polymerization reaction to displace 5′ pyrophosphate from the dNTP to be added. [The template strand is read in what direction?29] DNA pol is part of a large complex of proteins called the replisome. Other accessory proteins in this complex help DNA polymerase and allow it to polymerize DNA quickly. The prokaryotic replisome contains 13 components, and the eukaryotic replisome contains 27 proteins; additional complexity in the eukaryotic system is required because replication machinery must also unwind DNA from histone proteins.
Rapid elongation of the daughter strands follows. Since the two template strands are antiparallel, the two primers will elongate toward opposite ends of the chromosome. After a while it looks like this:
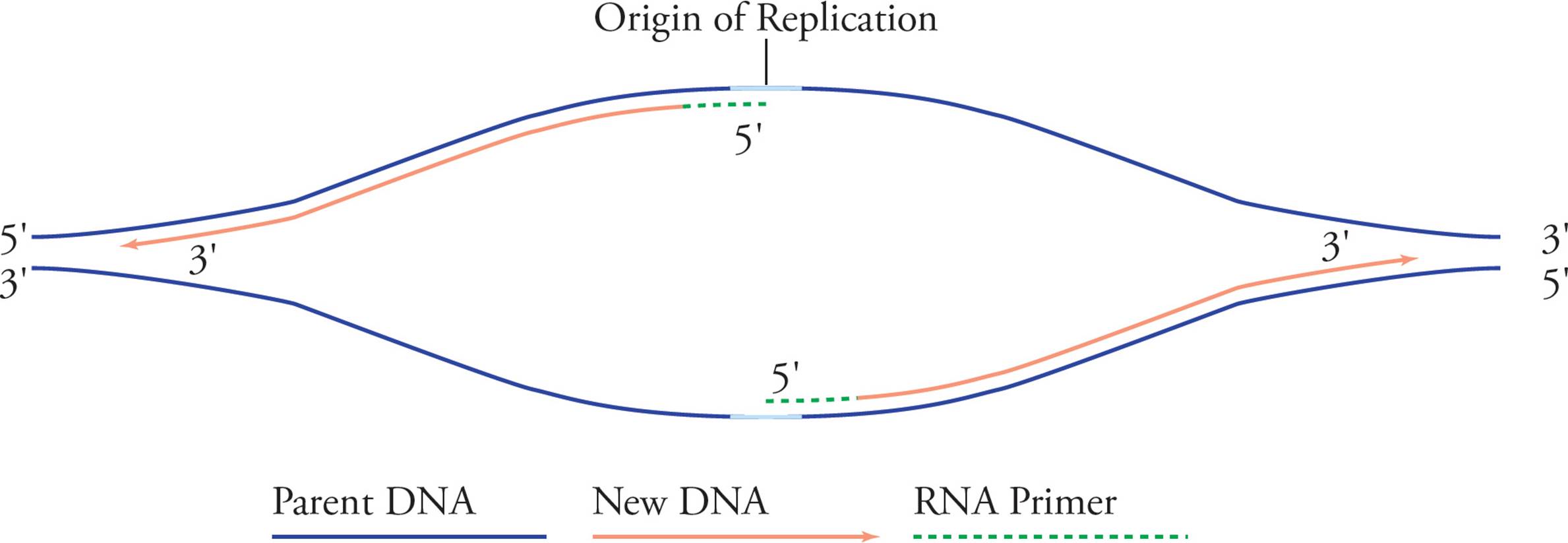
Figure 14 Elongation
DNA polymerase checks each new nucleotide to make sure it forms a correct base-pair before it is incorporated in the growing polymer. The thermodynamic driving force for the polymerization reaction is the removal and hydrolysis of pyrophosphate (P2O74–) from each dNTP added to the chain. (This is an example of a coupled reaction, discussed in Chapter 4.) Here are some more replication rules to memorize:
2) Polymerization occurs in the 5′ to 3′ direction, without exception. This means the existing chain is always lengthened by the addition of a nucleotide to the 3′ end of the chain. There is never 3′ to 5′ polymerase activity.
3) DNA pol requires a template. It cannot make a DNA chain from scratch but must copy an old chain. This makes sense because it would be pretty useless if DNA pol just made a strand of DNA randomly, without copying a template.
4) DNA pol requires a primer. It cannot start a new nucleotide chain.
• Can DNA polymerase make the following partially double-stranded structure completely double stranded in the presence of excess nucleotides, using the top strand as a primer?30
Replication proceeds along in both directions away from the origin of replication. Both template strands are read 3′ to 5′ while daughter strands are elongated 5′ to 3′. The areas where the parental double helix continues to unwind are called the replication forks. Let’s split the above picture (Figure 14) and look at an enlargement of the right side:
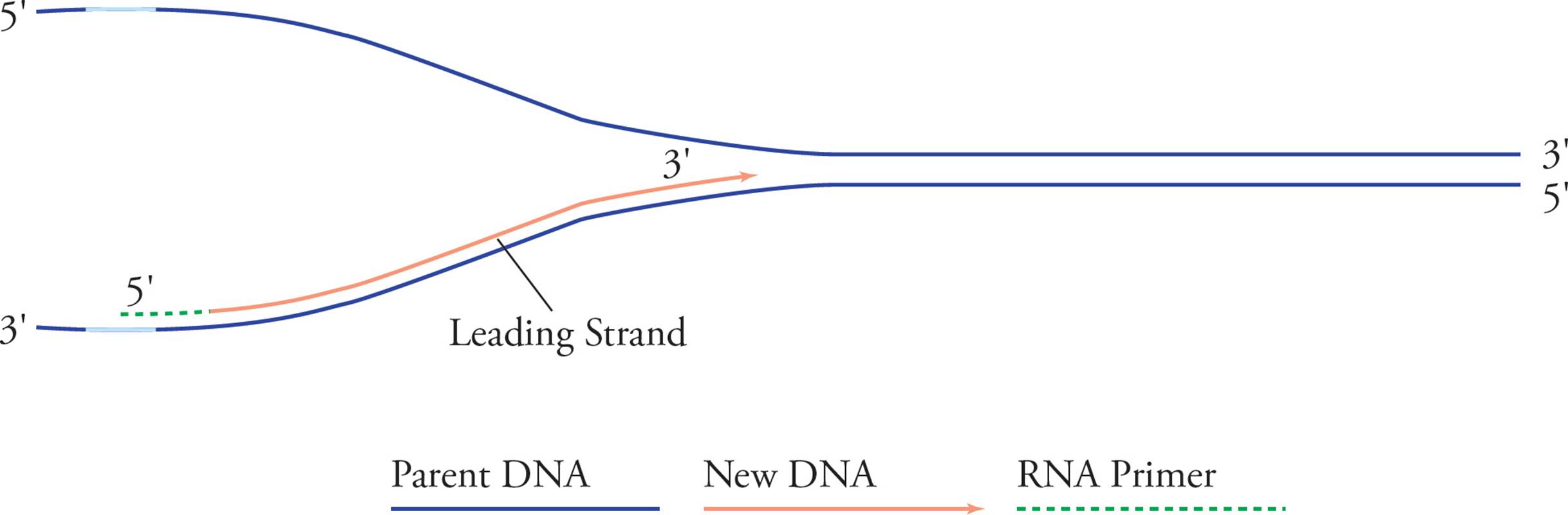
Figure 15 Leading Strand
See how it looks like a big fork? In examining these pictures, you have probably become aware of a problem. It seems like only half of each template strand will be replicated (in Figure 14, the right half of the bottom strand and the left half of the top strand). The problem is that chain elongation can only proceed in one direction, 5′ to 3′, but in order to replicate the right half of the top chain and the left half of the bottom one continuously, we would have to go in the opposite direction. Here’s the solution:
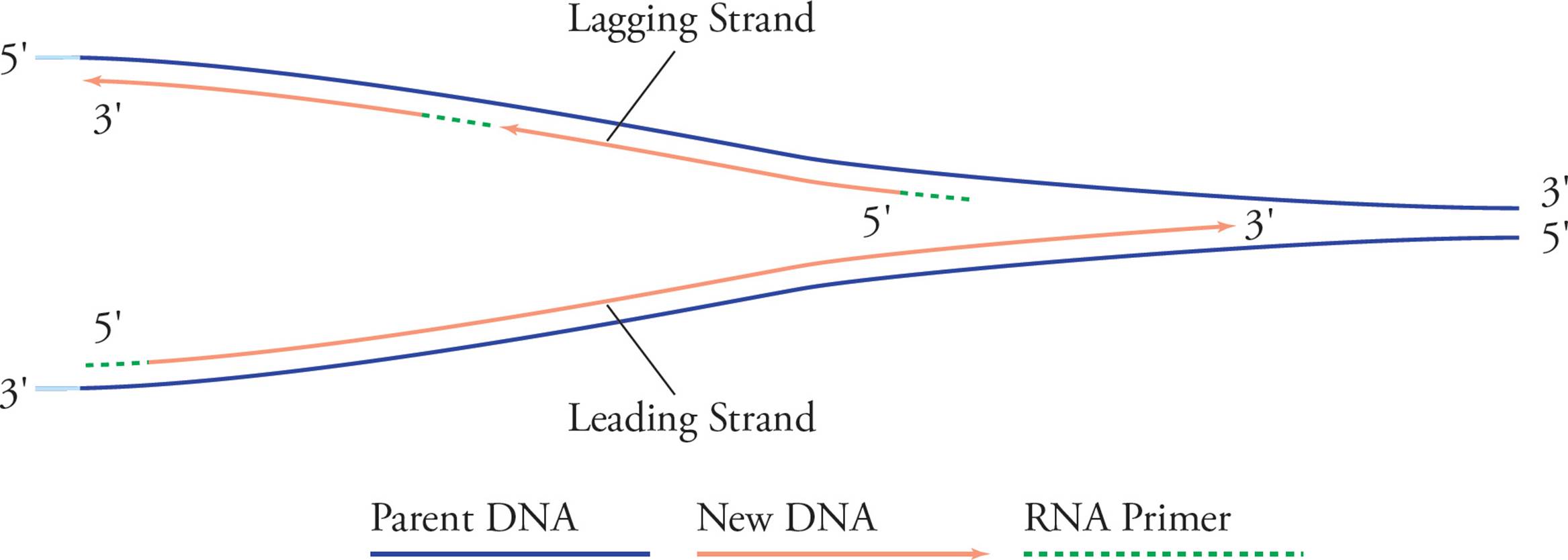
Figure 16 Leading and Lagging Strands
The solution to this problem involves building strands of DNA on opposite sides of the ORI using different methods. As the bottom chain on the right is elongated continuously, the replication fork widens. After a good bit of the top template chain becomes exposed, primase comes in and lays down a primer, which DNA pol can elongate. Then, when the replication fork widens again and more of the top template becomes exposed, these events are repeated. The bottom daughter on the right side, and the top daughter on the left side are called the leading strands because they elongate continuously right into the widening replication fork. The top daughter on the right, and the bottom daughter on the left are called the lagging strands because they must wait until the replication fork widens before beginning to polymerize. The small chunks of DNA comprising the lagging strand are called Okazaki fragments, after their discoverer. [As the replication forks grow, does helicase have to continue to unwind the double helix and separate the strands?31] Let’s continue our memory-list:
5) Replication forks grow away from the origin in both directions. Each replication fork contains a leading strand and a lagging strand.
6) Replication of the leading strand is continuous and leads into the replication fork, while replication of the lagging strand is discontinuous, resulting in Okazaki fragments.
7) Eventually all RNA primers are replaced by DNA, and the fragments are joined by an enzyme called DNA ligase.
DNA Polymerase
DNA polymerase can rapidly build DNA and is able to add tens of thousands of nucleotides before falling off the template. It is therefore said to be processive.
Eukaryotes have several different DNA polymerase enzymes, and their mechanisms of action are complex. You do not need to worry about this complexity.
Prokaryotes on the other hand have five types of DNA polymerases, called DNA polymerase I, II, III, IV and V. You should definitely know the functions of DNA pol III and DNA pol I:
1) DNA pol III is responsible for the super-fast, super-accurate elongation of the leading strand. In other words, it has high processivity. It has 5′ to 3′ polymerase activity as well as 3′ to 5′ exonuclease32 activity. This is when the enzyme moves backwards to chop off the nucleotide it just added, if it was incorrect; the ability to correct mistakes in this way is known as proofreading function. It has no known function in repair, and so is considered a replicative enzyme.
2) DNA pol I starts adding nucleotides at the RNA primer; this is 5′ to 3′ polymerase activity. Because of its poor processivity (it can only add 15-20 nucleotides per second), DNA pol III usually takes over about 400 base pairs downstream from the ORI. DNA pol I is also capable of 3′ to 5′ exonuclease activity (proofreading). DNA pol I removes the RNA primer via 5′ to 3′ exonuclease activity, while simultaneously leaving behind new DNA in __33activity. Finally, DNA pol I is important for excision repair (see below).
The functions of DNA pol II, IV, and V are less important to know for the MCAT:
3) DNA pol II has 5′ to 3′ polymerase activity, and 3′ to 5′ exonuclease proofreading function. It participates in DNA repair pathways and is used as a backup for DNA pol III.
4) DNA pol IV and DNA pol V have similar characteristics. They are error prone in 5′ to 3′ polymerase activity, but function to stall other polymerase enzymes at replication forks when DNA repair pathways have been activated. This is an important part of the prokaryotic checkpoint pathway. This enzyme has additional repair functions as well.
If a bacterium possesses a mutation in the gene for DNA polymerase III, resulting in an enzyme without the 3′ to 5′ exonuclease activity, will mutations occur more often than in bacteria with a normal DNA polymerase gene?34
Eukaryotic vs. Prokaryotic Replication
In eukaryotic replication, each chromosome has several origins. This is necessary because eukaryotic chromosomes are so huge that replicating them from a single origin would be too slow. As the many replication forks continue to widen, they create an appearance of bubbles along the DNA strand, so they are referred to as “replication bubbles.” Eventually the replication forks meet, and the many daughter strands are ligated together.
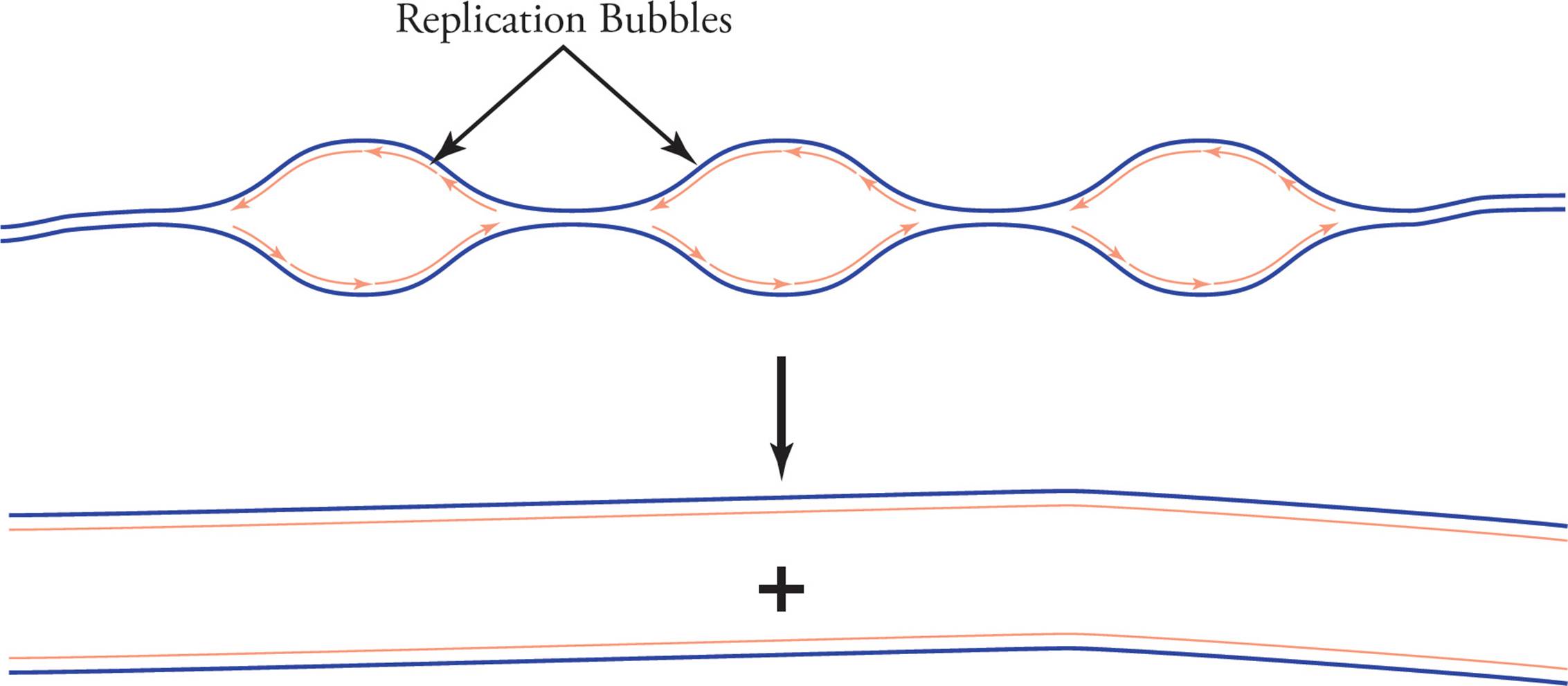
Figure 17 Eukaryotic Replication
Prokaryotes have only one chromosome, and this one chromosome has only one origin. Because the chromosome is circular, as replication proceeds the partially duplicated genome begins to look like the Greek letter θ (theta). Hence the replication of prokaryotes is said to proceed by the theta mechanism and is referred to as theta replication (see Figure 18).
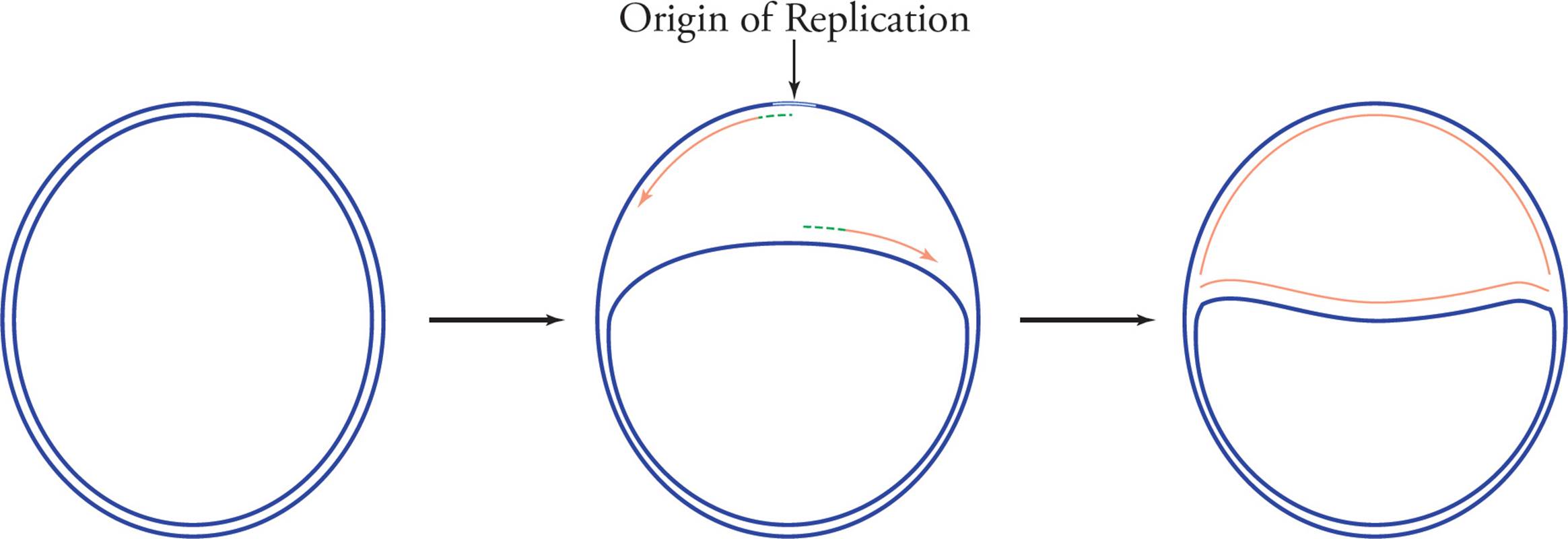
Figure 18 Theta (θ) Replication
Replicating Telomeres
DNA polymerase can only build DNA in one direction (5′ to 3′), and requires both a template and a primer. These requirements lead to a roadblock at chromosome ends. Eventually there will be no place on the lagging strand to lay down a primer, and primers close to the end of DNA cannot be replaced with DNA because there is nothing on the other side (DNA polymerase usually uses a previous length of upstream DNA to replace the primer, but this isn’t available at the end of a chromosome). This means that DNA replication machinery is unable to replicate sequences at the very ends of chromosomes, and after each round of the cell cycle and DNA replication, the ends of chromosomes shorten. Telomeres are disposable repeats at the end of chromosomes. They are consumed and shorten during cell division, becoming between 50 and 200 base pairs shorter.
When telomeres become too short, they reach a critical length where the chromosome can no longer replicate. As a consequence, cells can activate DNA repair pathways, enter a senescent state (where they are alive but not dividing), or activate apoptosis (pre-programmed cell death). The Hayflick limit is the number of times a normal human cell type can divide until telomere length stops cell division. Many age-related diseases are linked to telomere shortening.
Telomerase is an enzyme that adds repetitive nucleotide sequences to the ends of chromosomes and therefore lengthens telomeres. Telomerase is a ribonucleoprotein complex, containing an RNA primer and reverse transcriptase enzyme. Reverse transcriptases read RNA templates and generate DNA. In humans, the RNA template is 3′-CCCAATCCC-5′, and this allows for chromosome extension, one DNA repeat (5′-TTAGGG-3′) at a time (Figure 19). The telomerase complex continuously polymerizes, then translocates, allowing extension of six-nucleotide telomere repeats.
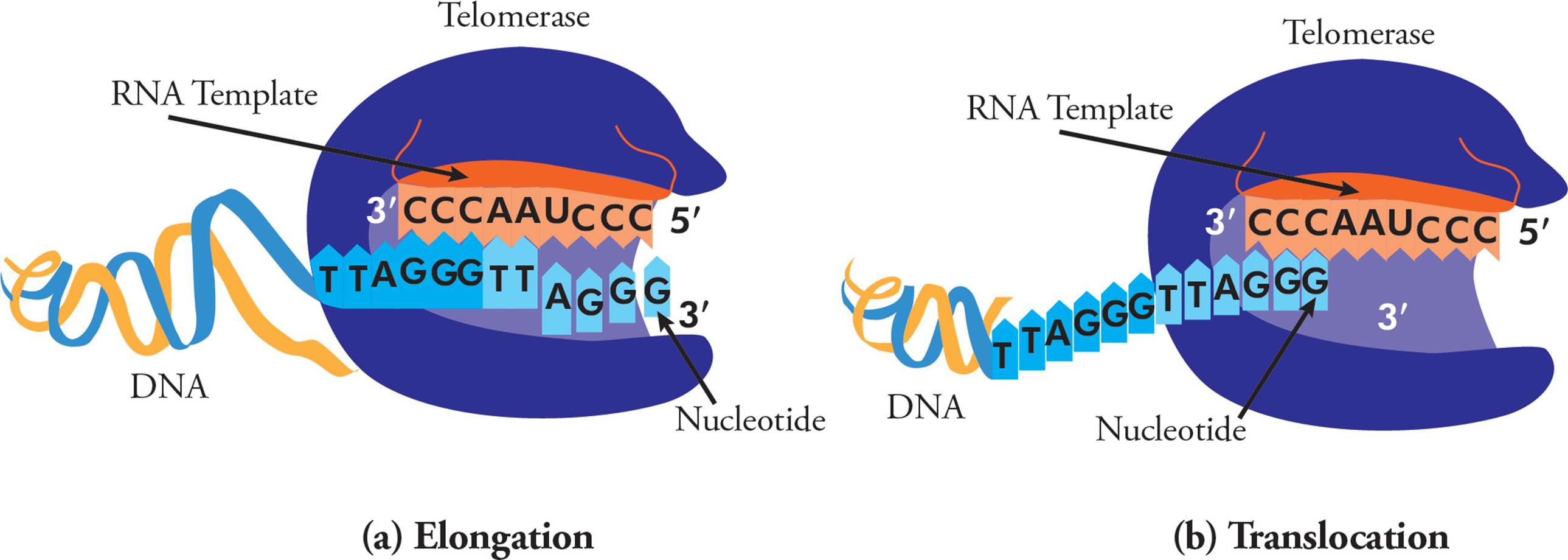
Figure 19 Telomerase and Telomere Lengthening
In most organisms, telomerase is only expressed in the germ line, embryonic stem cells, and some white blood cells. However, cancer cells can also express telomerase, which can help the cells immortalize. Telomere extension allows the cells to bypass senescence and apoptosis, and can therefore contribute to their transformation to a pre-cancerous state.
5.5 GENETIC MUTATION
Genetic mutation refers to any alteration of the DNA sequence of an organism’s genome. These can be inherited or acquired throughout life. Mutations that can be passed onto offspring are called germline mutations, since they occur in the germ cells (which give rise to gametes). Somatic mutations occur in somatic (non-gametic) cells and are not passed onto offspring. In other words, somatic mutations can have a major effect on an individual, but will not be passed on to future individuals in that population. Our cells have evolved elaborate repair pathways to help deal with mutations, and these will be discussed in the next section.
Causes of Mutation
There are many causes of mutation. Most are induced by an environmental factor or chemical, however they can also occur spontaneously.
Physical Mutagens
Ionizing radiation (such as X-rays, alpha particles and gamma rays) can cause DNA breaks. If these only occur on one strand (Figure 20, left), they can be easily patched up because the DNA helix is still held together in one piece. However, if both backbones are broken close to each other on a segment of DNA, a double-strand break (DSB) occurs (Figure 20, right). Here, the chromosome has been split into two pieces and it’s much more difficult to piece them back together.
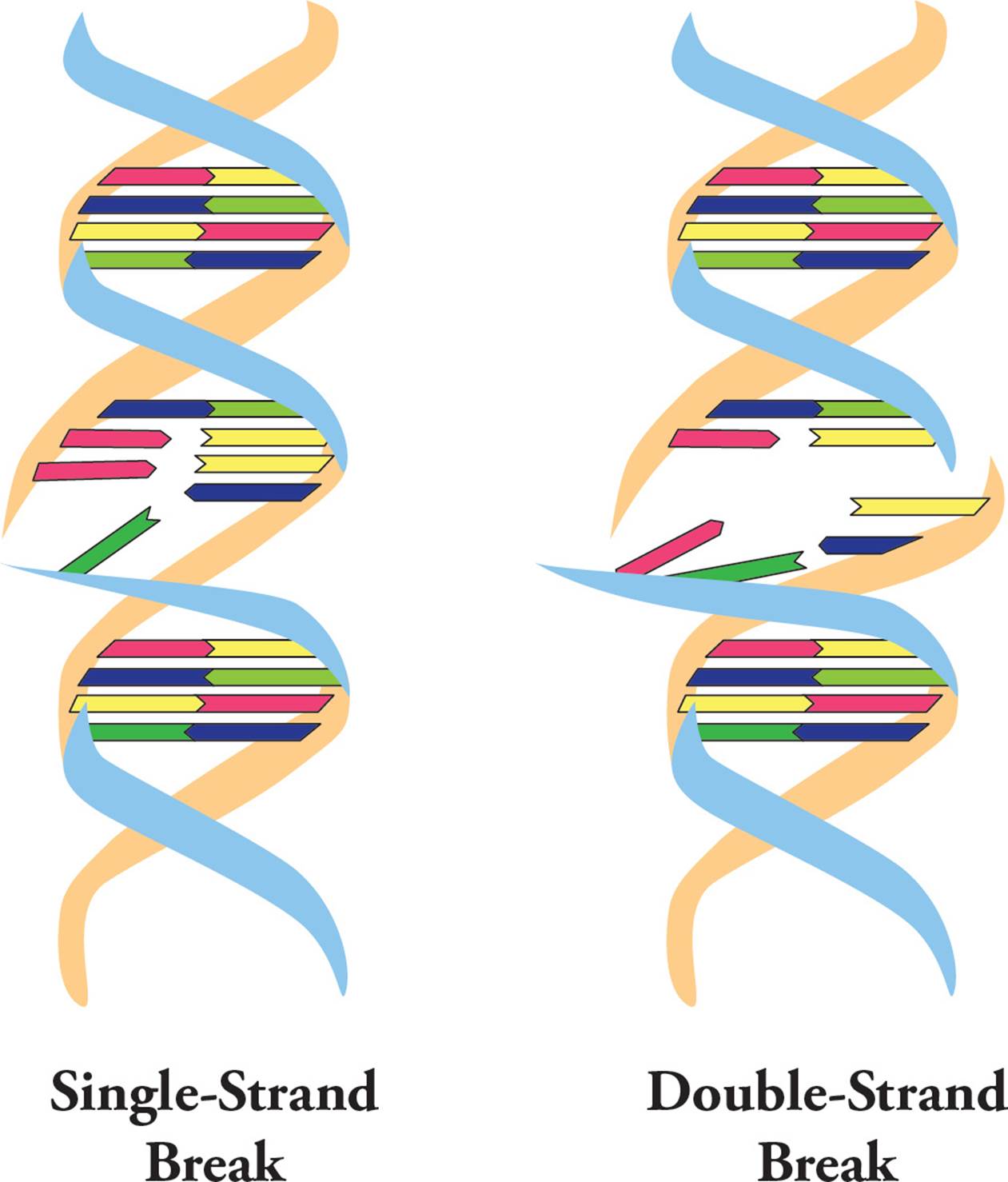
Figure 20 Single and Double Strand Breaks in DNA
UV light causes photochemical damage to DNA. For example, if two pyrimidines (two Cs or two Ts) are beside each other on a DNA backbone, UV light can cause them to become covalently linked. These pyrimidine dimers distort the DNA backbone (Figure 21), and can cause mutations during DNA replication if they are not repaired.
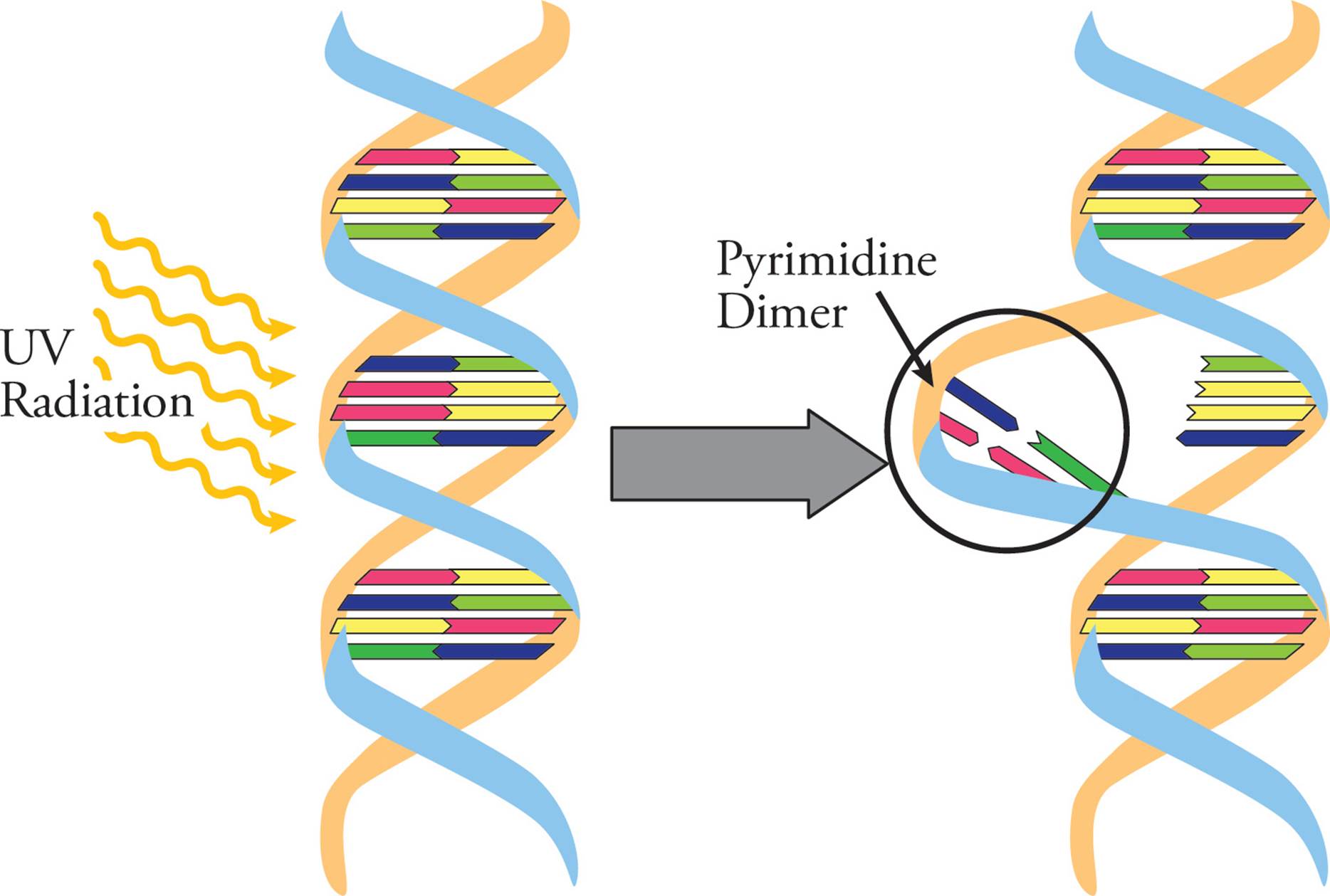
Figure 21 Pyrimidine Dimers in the DNA Helix
Reactive Chemicals
Many chemicals interact directly with DNA, and many others turn into damaging agents as they’re being processed by a cell. Chemicals can covalently alter bases, or can cause cross-linking or strand breaks. Cross-links are abnormal covalent bonds between different parts of DNA. Any compound that can cause mutations is called a mutagen.
Compounds that look like purines and pyrimidines (with large flat aromatic ring structures) cause mutations by inserting themselves between base pairs, or intercalating, thereby causing errors in DNA replication. Ethidium bromide is often used to visualize nucleic acids during gel electrophoresis in molecular biology labs (see Appendix I). This chemical is used because it is planar (and therefore intercalates with the DNA ladder), and glows orange when exposed to UV light (meaning nucleic acids in a gel can be easily visualized). However, because it intercalates with DNA, is also distorts the structure and can therefore disrupt DNA replication and transcription. Thus, ethidium bromide is a mutagen.
Biological Agents
Biological agents can also cause mutations. For example, although DNA polymerase has proofreading and correction abilities, it can still make a mistake. An incorrect base pair may be repaired (see Section 5.6), but if not, it will be passed on to all daughter cells. In this case, there is no mutagen. The mistake is spontaneous. Viruses can also affect DNA. Lysogenic viruses insert into the genome of the host cell (see Chapter 6), and this can cause mutations and disrupt genetic function. Some viruses can cause cancer because of this function. And finally, transposons can induce mutations. This will be described in the next section.
Types of Mutations
Based on structure, there are seven kinds of mutations:
1) Point mutations
2) Insertions
3) Deletions
4) Inversions
5) Amplifications
6) Translocations and rearrangements
7) Loss of heterozygosity
Point mutations are single base pair substitutions (A in place of G, for example). Point mutations can be transitions (substitution of a pyrimidine for another pyrimidine or substitution of a purine for another purine) or transversions(substitution of a purine for a pyrimidine or vice versa). There are three types of point mutations:
1) Missense mutation: causes one amino acid to be replaced with a different amino acid. This may not be serious if the amino acids are similar. [How can this occur?35]
2) Nonsense mutation: a stop codon replaces a regular codon and prematurely shortens the protein
3) Silent mutation: a codon is changed into a new codon for the same amino acid, so there is no change in the protein’s amino acid sequence
Insertion refers to the addition of one or more extra nucleotides into the DNA sequence, and deletion is the removal of nucleotides from the sequence. Both of these mutations can cause a shift in the reading frame. For example, AAACCCACC is read as AAA, CCC, ACC. It would code for Lys-Pro-Thr. Inserting an extra G into the first codon could produce this: AGAACCCACC. This would be read AGA, ACC, CAC, C. It now codes for Arg-Thr-His (plus there’s an extra C). Not only has the first codon and amino acid changed, the whole gene will be read differently and all amino acids in the protein from that point on will change. Mutations that cause a change in the reading frame are called frameshift mutations. Generally speaking, frameshift mutations are very serious. Note that a frameshift can lead to premature termination of translation (yielding an incomplete polypeptide) if it results in the presence of an abnormal stop codon. [Are all insertions and deletions frameshift mutations?36 If the following oligonucleotide is mutated by inserting a G between the fifth and sixth codons, what effect will this have on the oligopeptide it encodes: AUG AAG GGG CCC UUU AAA UGA CCC?37 For each type of mutation, does it involve a change in the genotype, the phenotype, or both?38]
In addition to mutations at individual nucleotides, larger-scale mutations are also common. Insertions and deletions can involve thousands of bases. An inversion is when a segment of a chromosome is reversed end to end. The chromosome undergoes breakage and rearrangement within itself (Figure 22).
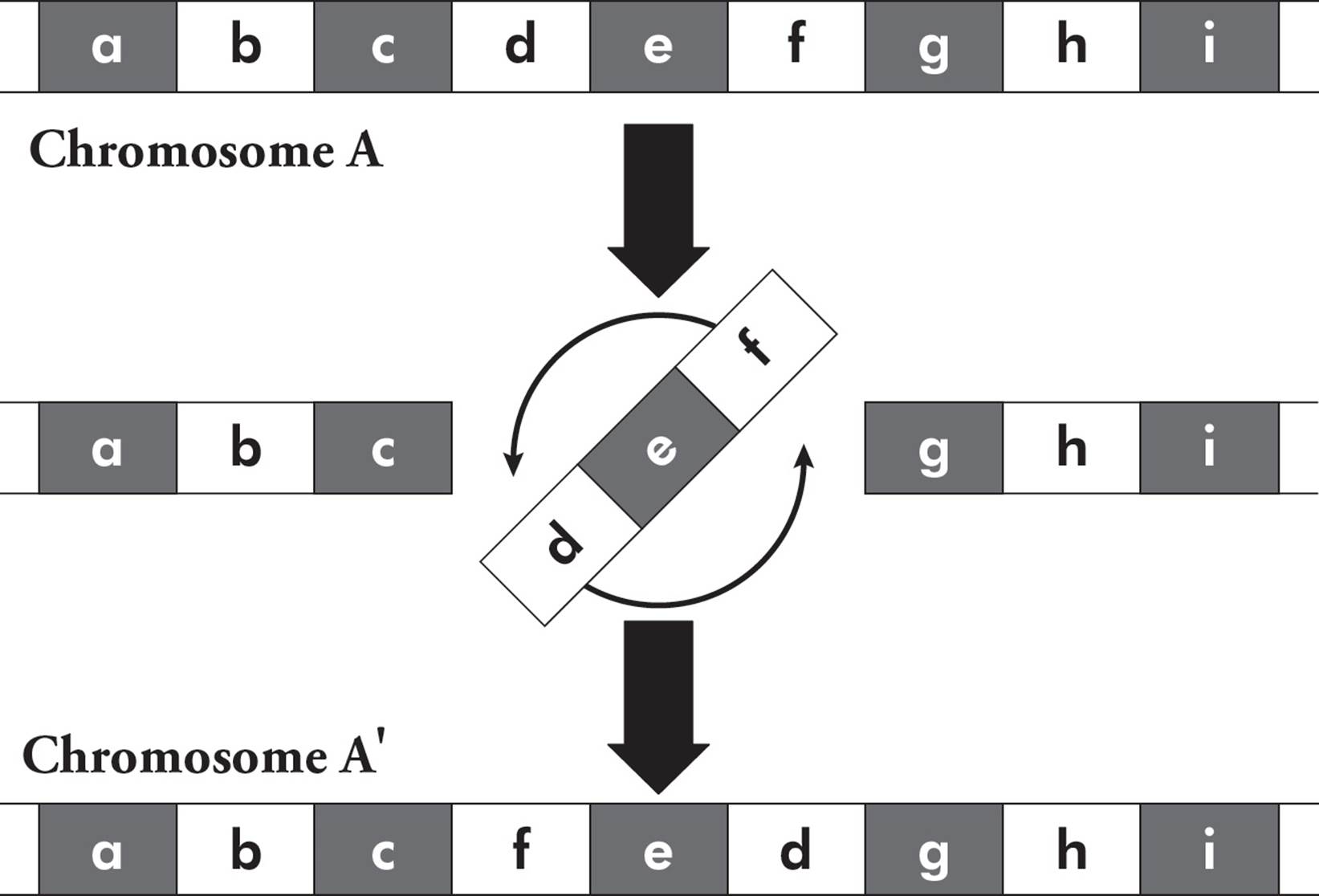
Figure 22 Chromosome Inversion
Chromosome amplification is when a segment of a chromosome is duplicated. This is similar to copy number variations discussed above. Translocations result when recombination occurs between nonhomologous chromosomes (Figure 23). This can create a gene fusion, where a new gene product is made from parts of two genes that were not previously connected. This is a common occurrence in many types of cancer. Translocations can be balanced (where no genetic information is lost), or unbalanced (where genetic information is lost or gained).

Figure 23 Chromosome Amplification
Transposons were introduced in Section 5.2. These mobile genetic sequences are commonly found in genomes and often cause mutations. When transposons are mobilized (Figure 10), they can insert in any part of the genome, and this can affect gene expression or cause mutations. They can jump into a promoter and turn gene expression off. They could jump into a protein-coding region and disrupt (or mutate) the sequence. They can also jump into regulatory parts of the genome and ramp up gene expression at a nearby site.
In addition to jumping around the genome, transposons can cause structural changes to chromosomes when they work in pairs. Directionality of the transposon is important here, as it determines what happens to the chromosome. If a chromosome has two transposons with the same direction (Figure 24), the transposons can line up beside each other, so they are parallel. This causes the chromosomal segment between them to loop around. Recombination occurs between the transposons, and this causes deletion of the DNA between the two transposons. The original chromosome therefore completely loses the DNA segment between the transposons (a deletion). The segment of DNA that is lost takes one transposon with it, meaning it can actually jump back into the genome somewhere else, causing chromosome rearrangement: one chunk of a chromosome has moved to a new location in the genome.
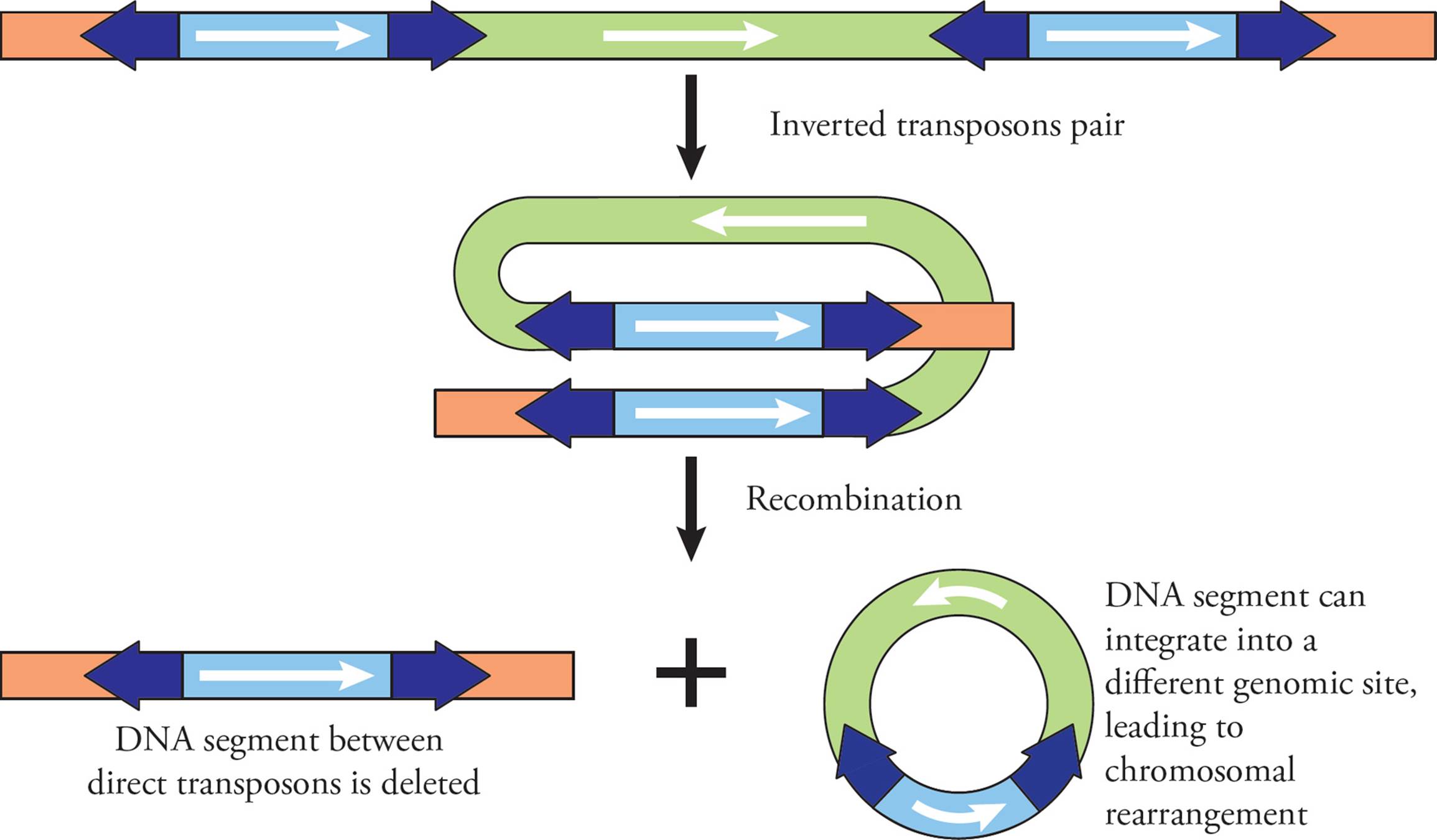
Figure 24 Deletion and Chromosomal Rearrangements via Transposons
If a chromosome has two transposons with inverted orientations (Figure 25), they can again pair and align with each other. After recombination, the sequence of DNA between the two transposons ends up inverted.
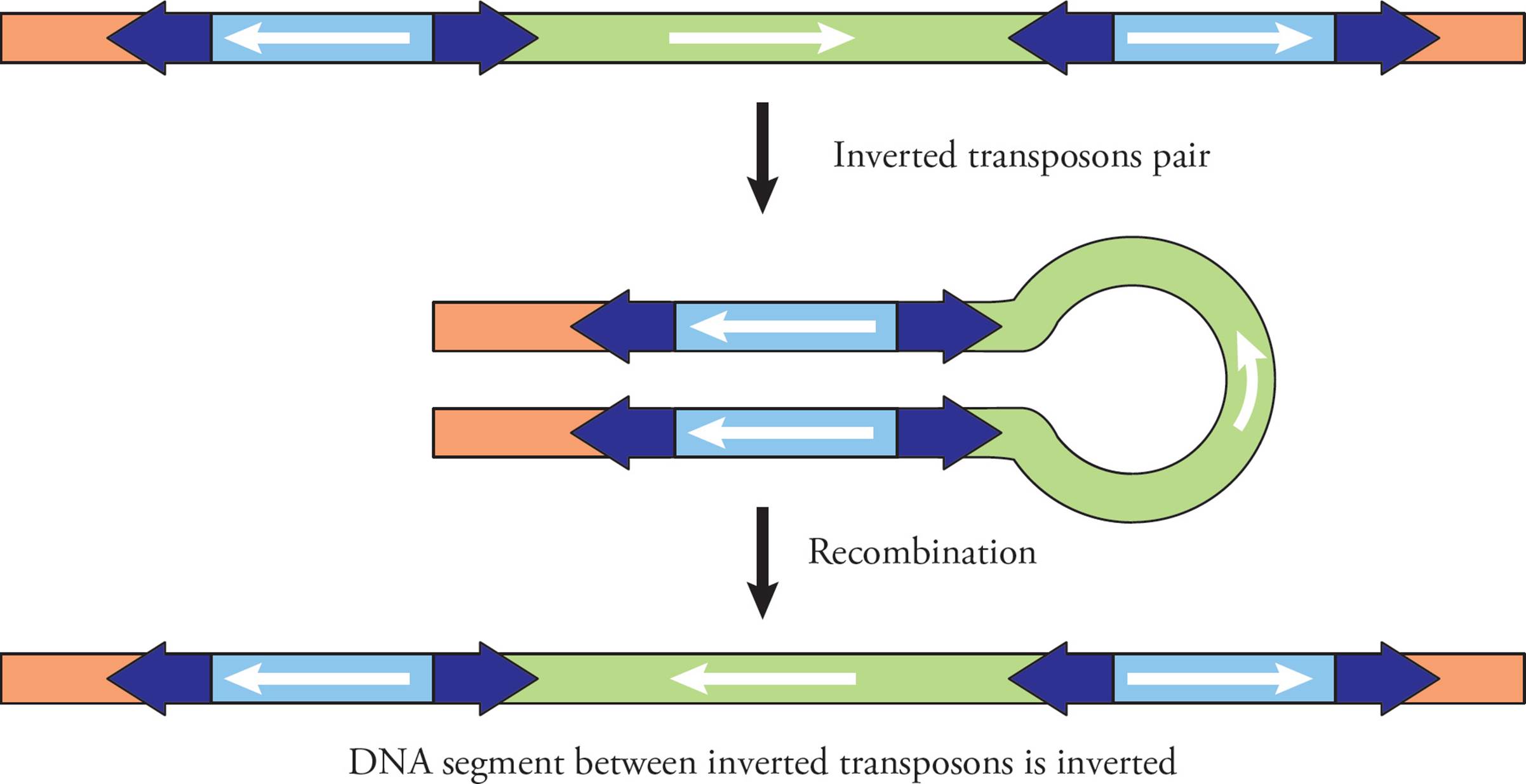
Figure 25 Chromosome Inversion via Transposons
Loss of heterozygosity occurs in a diploid organism when one allele of a certain gene is lost, either due to deletion or a recombination event. This makes the locus hemizygous: there is only one gene copy in a diploid organism. If the remaining allele is mutant or defective, all gene expression of this gene product is lost. For example, hereditary retinoblastoma is a type of retinal cancer common in young children. It occurs when a child receives a flawed copy of the tumor suppressor Rb1 from one parent. Loss of heterozygosity can lead to loss of the normal allele (from the other parent). With no functional Rb protein (due to having only one copy of Rb1, and it being a flawed or mutant copy), the child almost invariably develops retinoblastoma.
Effects of Mutations
There are many mechanisms by which mutations can exert their effects on the cell. A single amino acid change can affect protein activity, localization, degradation, half-life, or interactions, or, it may have no effect at all. The outcome of a mutation on a protein depends on where the mutation occurs. Mutations on sex chromosomes typically have a greater effect than mutations on autosomes since autosomes are present in double copies. Males have only one X chromosome and one Y chromosome, with no back-up copy of either. Similarly, most females only express one of their X chromosomes (see Section 5.9), and so they, too, often don’t have a back-up copy. Haploid expression in a diploid organism is hemizygosity, and this can lead to an increased effect of mutations on these chromosomes.
Gain-of-function mutations increase the activity of a certain gene product, or change it such that it gains a new and abnormal function. Loss-of-function mutations are the opposite; they result in the gene product having less or no function. In haploinsufficiency, a diploid organism has only a single functional copy of a gene, and this single copy is not enough to support a normal state. Haploinsufficiency highlights the importance of gene dose: many times, just expressing a gene is not enough. You must express enough of the gene to maintain good health.
Good and Bad Mutations
Despite the bad reputation they have, not all mutations are bad. Many mutations are neutral, and have no effect. Evolution is based on mutations and selection, and some mutations are beneficial. Those that confer a survival advantage will be selected for in a population.
There are examples of beneficial mutations in humans:
• Sickle-cell anemia is caused by mutations in the gene for hemoglobin (Hb). One of the most common mutations allows deoxygenated Hb to dimerize and form long chains, which distorts the red blood cell shape, causing it to sickle. These deformed cells cannot function properly and are prematurely destroyed, leading to anemia. However, people who carry this gene also have an advantage in that they are more resistant to malaria. In areas where malaria is common, this is an important benefit.
• Some humans are missing 32 base pairs in a gene called CCR5. This deletion confers HIV resistance to homozygotes and delays AIDS onset in heterozygotes. This mutation may have also conferred resistance to diseases in the past (such as the bubonic plague or smallpox), explaining its prevalence in populations of European descent, where these diseases were prevalent.
Mutations can also be disease causing. In some cases, one mutation is sufficient to induce a diseased state. In other cases, many mutations have to cooperate and occur together to cause a disease.
Inborn errors of metabolism are a huge group of genetic diseases that involve disorders of metabolism. Most of these are due to a single mutation in a single gene that codes for some sort of metabolic enzyme. Symptoms are caused by either the build-up of a toxic compound that can’t be broken down or by the deficiency of an essential molecule that cannot be synthesized. Because cellular metabolism is crucial, many symptoms are possible and a wide range of systems can be affected. Inborn errors of metabolism are typically organized into groups of disorders, depending on what type of metabolic pathways they affect: carbohydrate, amino acid, urea cycle, organic acids, fatty acid oxidation, mitochondrial, porphyrin, purine or pyrimidine, steroid, peroxisomal function, or lysosomal storage.
Cancer is driven by mutation accumulation. These mutations can either be inherited, or can be caused by carcinogen exposure. A carcinogen is a mutagen that is directly involved in causing cancer. Tumors typically have hundreds of mutations, ranging from point mutations to massive chromosomal changes. These mutations are often in oncogenes and tumor suppressors. An oncogene is a gene that can cause cancer when it is mutated or expressed at high levels. Tumor suppressors are the opposite, in that their deletion (or expression at decreased levels) can cause cancer. Some mutations will drive tumor growth and are highly selected for. These mutations are the most promising targets for developing cancer treatments, as the cancer cells rely on these mutations for growth.
5.6 DNA REPAIR
Cells have developed several mechanisms to deal with DNA damage. First, cell cycle checkpoints are activated, and arrest cell cycle progression. In eukaryotes, checkpoint pathways function at phase boundaries (such as the G1/S transition, and the G2/M transition), and can also be activated within some phases. Extensive DNA damage can induce apoptosis in eukaryotes, but before this happens, cells try to repair the DNA damage. This is important so that defective DNA isn’t passed on to daughter cells. There are several types of DNA repair.
Direct Reversal
Many types of DNA damage are irreversible and require repair pathways to fix the damage. However, a few can be directly reversed. For example, some enzymes can repair UV-induced pyrimidine photodimers using visible light. This process is called photoreactivation, and directly repairs the UV damage to DNA. This is commonly performed by bacteria and many plants. If pyrimidine dimers are not directly repaired, nucleotide excision repair can be used instead (see below). This is the main mechanism of repair in humans, but can introduce a mutation when trying to complete the repair. If left unrepaired, pyrimidine dimers in humans may lead to melanoma, a type of very dangerous and malignant skin tumor.
Homology-Dependent Repair
One of the benefits of DNA structure is the presence of a back-up copy; because DNA is double stranded, mutations on one strand of DNA can be repaired using the undamaged, complementary information on the other strand. Repair pathways that rely on this characteristic of DNA are called homology-dependent repair pathways. These can be divided into repair that happens before DNA replication (excision repair), or repair that happens during and after DNA replication (post-replication repair).
Excision Repair
Excision repair involves removing defective bases or nucleotides and replacing them. If these bases are not repaired, they can induce mutations during DNA replication, since replication machinery cannot pair them properly.
Post-Replication Repair
The mismatch repair pathway (MMR) targets mismatched Watson-Crick base pairs that were not repaired by DNA polymerase proofreading during replication. To do this, mispaired bases must be identified and fixed, but the crucial question is: which base is the correct one and which is the mistake? For example, if DNA contains an AC base pair, is the adenine correct and C should be removed and replaced with T? Or is the cytosine correct and A should be removed and replaced with G?
Some bacteria use genome methylation to help differentiate between the older DNA template strand and the newly synthesized daughter strand. Methylation takes a while to complete, which means that shortly after DNA synthesis, the parental template strand will be labeled with methylated bases and the new daughter strand will not. Bacterial machinery can read these methyl tags and know which base is the correct one (the one on the older strand) and which needs to be replaced (the newer one).
Other prokaryotes and most eukaryotes use a different system, where the newly synthesized strand is recognized by the free 3′-terminus on the leading strand, or by the presence of gaps between Okazaki fragments on the lagging strand.
Double-Strand Break Repair
DNA double-strand breaks (DSBs) can be caused by reactive oxygen species, ionizing radiation, UV light or chemical agents. Cells have two pathways to help in DSB repair: homologous recombination and nonhomologous end-joining. The goal of both is to reattach and fuse chromosomes that have come apart because of DSB. If done incorrectly, this can lead to deletions (where genetic information is lost) or translocations (where chromosome segments move to other chromosomes).
Homologous Recombination
After DNA replication, the genome contains identical sister chromatids. Homologous recombination is a process where one sister chromatid can help repair a DSB in the other. First, the DSB is identified and trimmed at 5′ ends to generate single-stranded DNA (Figure 26). This is done by nucleases (which break phosphodiester bonds) and helicase (to unwind the DNA). Many proteins bind these ends and start a search of the genome to find a sister chromatid region that is complementary to the single-stranded DNA. Once found, the complementary sequences are used as a template to repair and connect the broken chromatid. This requires a “joint molecule,” where damaged and undamaged sister chromatids cross over. DNA polymerase and ligase build a corrected DNA strand.
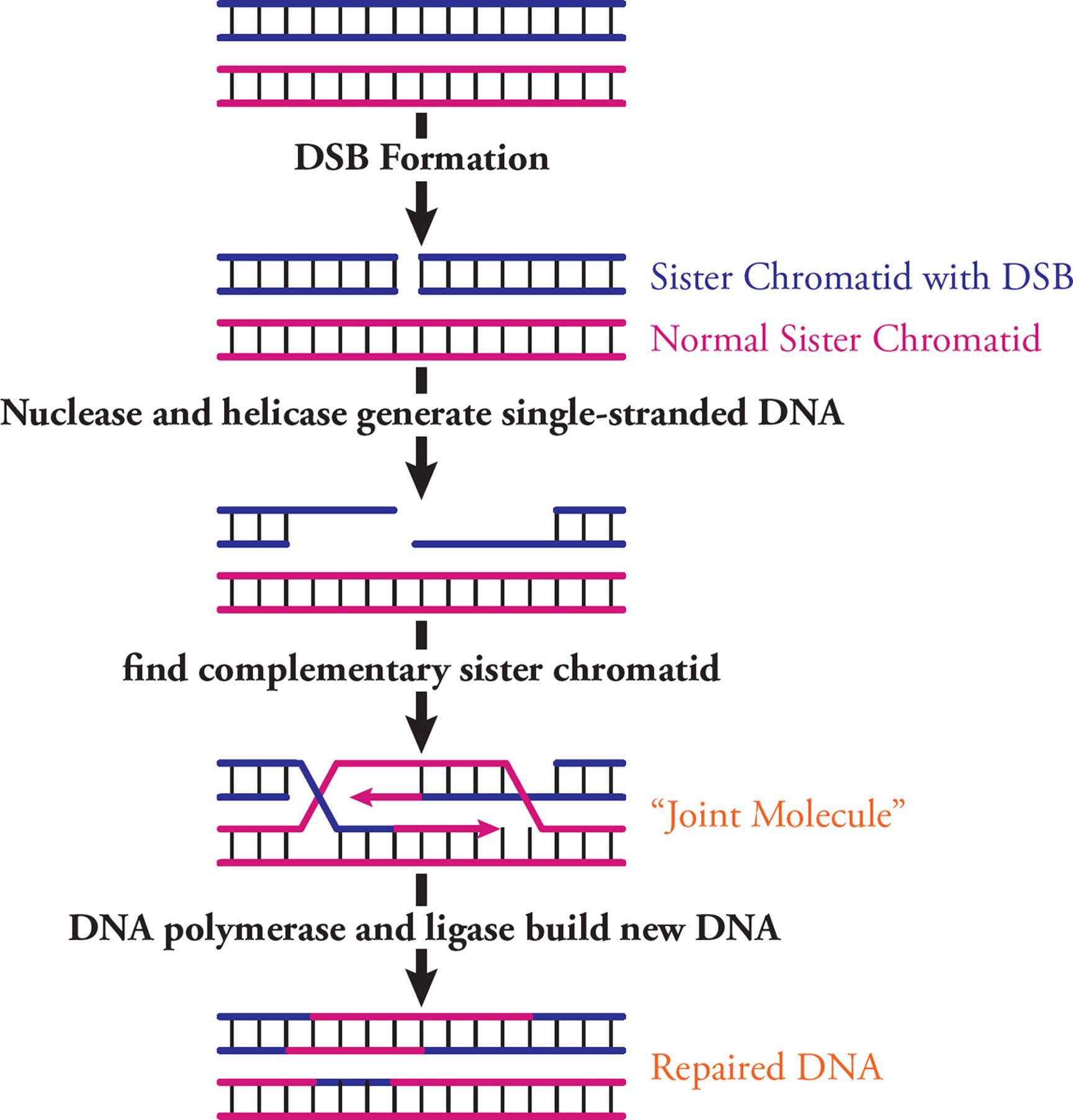
Figure 26 Homologous Recombination to Repair Double-Strand Breaks
Nonhomologous End Joining
Cells that aren’t actively growing or cycling through the cell cycle don’t have the option of using sister chromatids to repair DSBs in an error-free way. Since DNA replication isn’t happening, there is no chromosome backup to use. In this case, even a poorly repaired chromosome is better than one with a DSB, since chromosome breaks can lead to rearrangements.
Nonhomologous end joining is used to accomplish repair in this case. This process is common in eukaryotes but relatively uncommon in prokaryotes. First, broken ends are stabilized and processed, then DNA ligase connects the fragments. Nothing about this process requires specificity; the goal is just to reconnect broken chromosomes. Often, this can result in base pairs being lost, or chromosomes being connected in an abnormal way.
5.7 GENE EXPRESSION: TRANSCRIPTION
Gene expression refers to the process whereby the information contained in genes begins to have effects in the cell. The Central Dogma tells us that genetic information must be written in the form of RNA (i.e., it must be transcribed); and then it must be expressed as protein (i.e., it must be translated). Hence, the logical place to begin our discussion of gene expression is with the nature of RNA and transcription.
Characteristics of RNA
RNA is chemically distinct from DNA in three important ways:
1) RNA is single-stranded, except in some viruses.
2) RNA contains uracil instead of thymine.
3) The pentose ring in RNA is ribose rather than 2′ deoxyribose.
As a result of this last difference, the RNA polymer is less stable, because the 2′ hydroxyl can nucleophilically attack the backbone phosphate group of an RNA chain, causing hydrolysis when the remainder of the chain acts as leaving group. This cannot occur in DNA, since there is no 2 hydroxyl. [Why is the stability of RNA relatively unimportant?39 Anticancer drugs often seek to block growth of rapidly dividing cells by inhibiting production of thymine. Why is this an attractive target for cancer therapy?40] This chemical property has a big impact in molecular biology labs, where DNA samples are stable at a range of temperatures for a relatively long period of time, but high quality RNA is difficult to extract and is only stable for a short time.
There are several different types of RNA, each with a unique role.
Coding RNA
You are already familiar with messenger RNA (mRNA), the only type of coding RNA. This molecule carries genetic information to the ribosome, where it can be translated into protein; each unique polypeptide is created according to the sequence of codons on a particular piece of mRNA, which was transcribed from a particular gene. To allow for this, each mRNA has several regions. The 5′ region is not translated into protein (so is called the 5′ untranslated region, or 5′UTR), but is important in initiation and regulation. Following the 5′UTR is the region that codes for a protein. This starts at a start codon and ends at a stop codon, and is called the open reading frame (ORF). The 3′ end of the mRNA (after the stop codon) isn’t translated into protein, but often contains regulatory regions that influence post-transcriptional gene expression (see Section 5.9).
Eukaryotic mRNA is usually monocistronic and obeys the “one gene, one protein” principle. This means that each piece of mRNA encodes only one polypeptide (and so contains one ORF). Hence, there are as many different mRNAs as there are proteins. Because each mRNA can be read many times, each transcript can be used to make many copies of its polypeptide. There are a few exceptions to the “one gene, one protein” principle; recently, some polycistronic eukaryotic mRNAs have been discovered, and these will be discussed below.
In contrast, prokaryotic mRNA often codes for more than one polypeptide and is termed polycistronic. Different open reading frames on the same polycistronic mRNA are generally related in function.41 Translation termination and initiation sequences are found between the ORFs. The termination information helps finish the previous peptide chain, and initiation information helps start translation of the next open reading frame on the transcript.
Messenger RNA is constantly produced and degraded, according to the cell’s need for the protein encoded by each piece of mRNA. In fact, this is the principal means whereby cells regulate the amount of each particular protein they synthesize. This is an important point that will be emphasized later. Note that in eukaryotes, the first RNA transcribed from DNA is an immature or precursor to mRNA called heterogeneous nuclear RNA (hnRNA). Processing events (such as addition of a cap and tail, and splicing) are required for hnRNA to become mature mRNA. Since prokaryotes do not process their primary transcripts, hnRNA is only found in eukaryotes.
Non-Coding RNA
Non-coding RNA (ncRNA) is a functional RNA that is not translated into a protein. The human genome codes for thousands of ncRNAs, and there are several types. The two major types to know for the MCAT are transfer RNA (tRNA) and ribosomal RNA (rRNA).
Transfer RNA (tRNA) is responsible for translating the genetic code. Transfer RNA carries amino acids from the cytoplasm to the ribosome to be added to a growing protein. The structure of tRNA and how it does its job will be discussed in Section 5.8. [Estimate how many different tRNAs there are.42]
Ribosomal RNA (rRNA) is the major component of the ribosome. Humans have only four different types of rRNA molecules (18S, 5.8S, 28S and 5S), although almost all the RNA made in a given cell is rRNA. All rRNAs serve as components of the ribosome, along with many polypeptide chains. One rRNA provides the catalytic function of the ribosome, which is a little odd. In most other cases, enzymes are made from polypeptides. Catalytic RNAs are also called ribozymes (or ribonucleic acid enzymes), since they are capable of performing specific biochemical reactions, similar to protein enzymes. There are additional examples of ribozymes, including snRNA (discussed below) and some introns that are self-splicing.
Some other interesting non-coding RNAs are:
• Small nuclear RNA (snRNA) molecules (150 nucleotides) associate with proteins to form snRNP (small nuclear ribonucleic particles) complexes in the spliceosome.
• MicroRNA (miRNA) and small interfering RNA (siRNA) function in RNA interference (RNAi), a form of post-transcriptional regulation of gene expression. Both can bind specific mRNA molecules to either increase or decrease translation. This will be discussed more in Section 5.9.
• PIWI-interacting RNAs (piRNAs) are single stranded and short (typically between 21 and 31 nucleotides in length). They work with a class of regulatory proteins called PIWI proteins to prevent transposons from mobilizing.
• Long ncRNAs are longer than 200 nucleotides. They help control the basal transcription level in a cell by regulating initiation complex assembly on promoters. They also contribute to many types of post-transcriptional regulation, by controlling splicing and translation, and they function in imprinting and X-chromosome inactivation (see Section 5.9).
Replication vs. Transcription
Transcription is the synthesis of RNA (usually mRNA, tRNA, or rRNA) using DNA as the template. The word transcription indicates that in the process of reading and writing information, the language does not change. Information is transferred from one polynucleotide to another. This should lead you to expect transcription to be fairly similar to replication. And it is.
Both replication and transcription involve template-driven polymerization. [Because of this, the RNA transcript produced in transcription is __43 to the DNA template, just as the daughter strand produced in replication was.] The driving force for both processes is the removal and subsequent hydrolysis of pyrophosphate from each nucleotide added to the chain, with the existing chain acting as nucleophile. [Transcription, like replication, can occur only in the __44 direction. Do the polymerase enzymes in both replication and transcription require a primer?45] Another important difference between transcription and DNA replication is that RNA polymerase has not been shown to possess the ability to remove mismatched nucleotides (it lacks exonuclease activity); in other words, it cannot correct its errors. Thus, transcription is a lower fidelity process than replication. [A virus possessing an RNA genome relies on RNA polymerase rather than DNA polymerase to replicate its genome. Will this virus have a higher or a lower rate of spontaneous mutation than organisms with ds-DNA genomes?46]
Another similarity is that transcription, like replication, begins at a specific spot on the chromosome. The name of the site where transcription starts (the start site) is different from the name of the place where replication begins, __.47 The sequence of nucleotides on a chromosome that activates RNA polymerase to begin the process of transcription is called the promoter, and the point where RNA polymerization actually starts is called the start site. In fact, from this point forward, just about every event in transcription is given a different name from the events in replication.
Reference Points in Transcription
Before we discuss the mechanics of transcription, we need to clarify a few reference points (see Figure 27). We noted previously that the chromosome is referred to as the template, not parent. What about the individual strands of the chromosome? Are they both templates for the same mRNA? Let’s answer with a thought experiment. Say there is a strand of DNA which has the sequence AAAAAAAAA. If we transcribe this strand, the resulting mRNA will look like: UUUUUUUUU. When it is translated, this mRNA will result in an oligopeptide with this primary structure: Phe-Phe-Phe. (Refer to the genetic code Table in Section 5.3.) Now, what if we transcribe the other strand of the chromosome? What is its DNA sequence? What will the transcript look like? And the oligopeptide?48 Our conclusion is that only one of the strands of the DNA template encodes a particular mRNA molecule. But it makes sense: paired DNA strands are complementary, not identical. The strand which is actually transcribed is called the template, non-coding, transcribed, or antisense strand; it is complementary to the transcript. The other DNA strand is called the coding or sense strand; it has the same sequence as the transcript (except it has T in place of U). It is customary to say that transcription starts at a point and proceeds downstream, which means toward the 3 end of the coding strand and transcript. Upstream means toward the 5 end of the coding strand, beyond the 5 end of the transcript. Upstream nucleotide sequences are referred to using negative numbers, and downstream sequences are referred to using positive numbers. The first nucleotide on the template strand which is actually transcribed is called the start site. The corresponding nucleotide on the coding strand is given the number +1. As we’ll see below, regulatory sequences on the chromosome are referred to by where they occur on the coding strand.
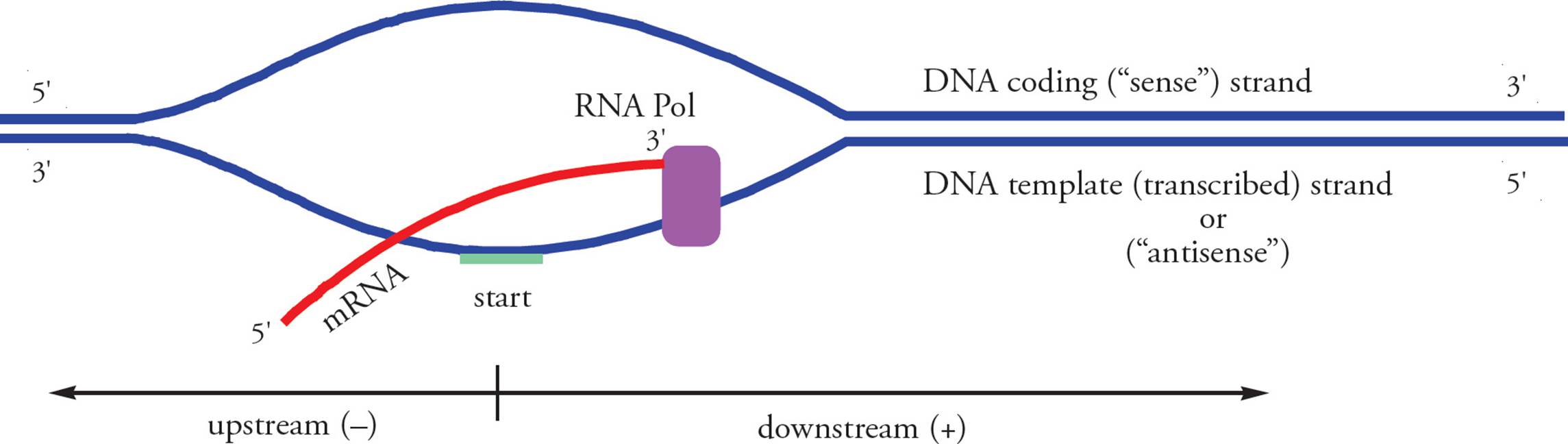
Figure 27 Reference Points in Transcription
• The Figure above labels the transcript “mRNA.” Is this accurate in all life forms? (Hint: In eukaryotes, is the initial transcript mature mRNA, ready to be translated?)49
Prokaryotic Transcription
It is important to understand all the vocabulary and general principles presented above. In this section and the next, we will present some more detailed information.
In bacteria (prokaryotes), all types of RNA are made by the same RNA polymerase. Prokaryotic RNA polymerase is a large enzyme complex consisting of five subunits: two alpha subunits, a beta subunit, a beta′ subunit, and an omega subunit (α2ββ'ω). This is the core enzyme responsible for rapid elongation of the transcript. However, the core enzyme alone cannot initiate transcription. An additional subunit termed the sigma factor (σ) is required to form what is sometimes referred to as the holoenzyme (holo = complete), which is responsible for initiation.
Transcription occurs in three stages: initiation, elongation, and termination. Initiation occurs when RNA polymerase holoenzyme binds to a promoter. The typical bacterial promoter contains two primary sequences: the Pribnow box at –10 and the –35 sequence. Holoenzyme scans along the chromosome like a train on a railroad track until it recognizes a promoter and then stops, forming a closed complex. The RNA polymerase must unwind a portion of the DNA double helix before it can begin to synthesize RNA. The RNA polymerase bound at the promoter with a region of single-stranded DNA is termed the open complex. Once the open complex has formed, transcription can begin.
The sigma factor plays two roles in helping the polymerase find promoters. The first is to greatly increase the ability of RNA polymerase to recognize promoters. The second is to decrease the nonspecific affinity of holoenzyme for DNA. Once the open complex and several phosphodiester bonds have been formed, the sigma factor is no longer necessary and leaves the RNA polymerase complex.
The core enzyme elongates the RNA chain processively, with one polymerase complex synthesizing an entire RNA molecule. As the core enzyme elongates the RNA, it moves along the DNA downstream in a transcription bubblein which a region of the DNA double helix is unwound to allow the polymerase to access the complementary DNA template. When a termination signal is detected, in some cases with the help of a protein called rho, the polymerase falls off of the DNA, releases the RNA, and the transcription bubble closes.
Comparing Prokaryotic and Eukaryotic Transcription
Eukaryotic and prokaryotic transcription are similar, but you need to be aware of four major differences. Differences in location, RNA polymerases and primary transcripts are discussed here. Regulation of transcription is another major difference and is discussed in Section 5.9.
Location
Eukaryotic means “true-kernelled.” Prokaryotic means “before-the-kernel.” The karyon (kernel) is, of course, the nucleus. The fact that prokaryotes have no nucleus means transcription occurs free in the cytoplasm, in the same compartment where translation occurs, and transcription and translation can occur simultaneously. Eukaryotes must transcribe their mRNA in the nucleus, then modify it (see below), then transport it across the nuclear membrane to the cytoplasm where it can be translated. Transcription and translation in eukaryotes do not occur simultaneously.
Another important difference between prokaryotic and eukaryotic gene expression is that the primary transcript in prokaryotes is mRNA. In other words, the product of transcription by prokaryotic RNA polymerase is ready to be translated. In fact, translation of prokaryotic mRNA begins before transcription is completed!
In contrast, the eukaryotic primary transcript (hnRNA made by RNA pol II, see below for info on eukaryotic RNA polymerases) is modified extensively before translation (Figure 30). The most important example is splicing. Eukaryotic DNA has non-coding sequences intervening between the segments that actually code for proteins. Sometimes these intervening sequences contain enhancers or other regulatory sequences and they can be quite long. The average size of a mammalian intron, for example, is about 2000 nucleotides. Intervening sequences in the RNA are called introns. Note that introns are intragenic regions (and not intergenic space, discussed in Section 5.2). Protein-coding regions of the RNA are termed exons because they actually get expressed. Before the RNA can be translated, introns must be removed and exons joined together; this is accomplished via splicing.
Splicing is mediated by the spliceosome, a complex that contains over 100 proteins and 5 small nuclear RNA (snRNA) molecules. About half the proteins stably bind snRNAs, and these form three small nuclear ribonucleic particles (snRNPs). Each snRNP is therefore made of proteins and snRNAs. The spliceosome is not a pre-assembled complex, but rather assembles around each intron that needs to be removed. This happens in a series of steps, where different snRNP components are recruited and released as the reaction proceeds. The complex undergoes many conformational changes to attain catalytic activity.
To catalyze the splicing reaction, snRNPs recognize and hydrogen bond to conserved nucleotides in the intron: typically GU at the 5′ end, AG at the 3′ end, and an adenine 15-45 bases upstream of the 3′ splice site. This aligns the hnRNA such that the splicing mechanism can take place (Figure 28). Two splicing reactions are catalyzed by the spliceosome. The first reaction attaches one end of the intron to the conserved adenine. This causes the intron to form a looped structure, then the second reaction joins the two exons (Figure 28) and releases the loop. The five conserved nucleotides necessary for this reaction (GU, A and AG) are found in all genes and across all eukaryotic species.
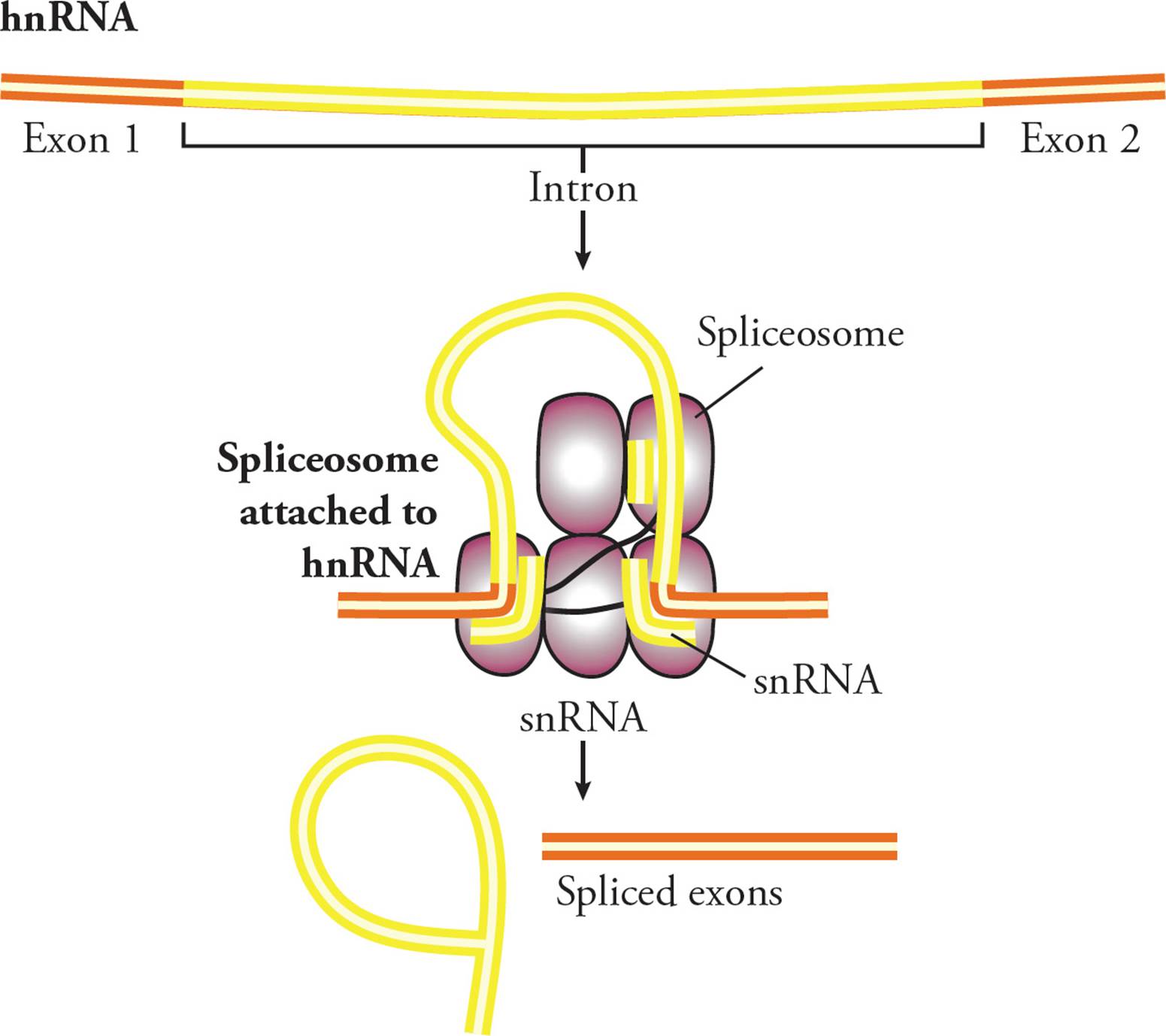
Figure 28 Mechanism of Splicing
For a given gene, there are often different options or patterns of splicing, a phenomenon called alternative splicing. There are many different common patterns. One gene could have different promoters in the 5′ region, which can change where/how the RNA begins. There can be alternative 5′ exons or 3′ exons, which can affect either end of the RNA. In the middle too, some exons can be included or skipped. Finally, there could be mutually exclusive exons, where sometimes one is included and sometimes the other is kept. All these patterns lead to different mRNAs being made from one DNA gene sequence; the mRNAs can be different in length and sequence. Shuffling exons in this way is one way to increase the complexity of gene expression (Figure 29).
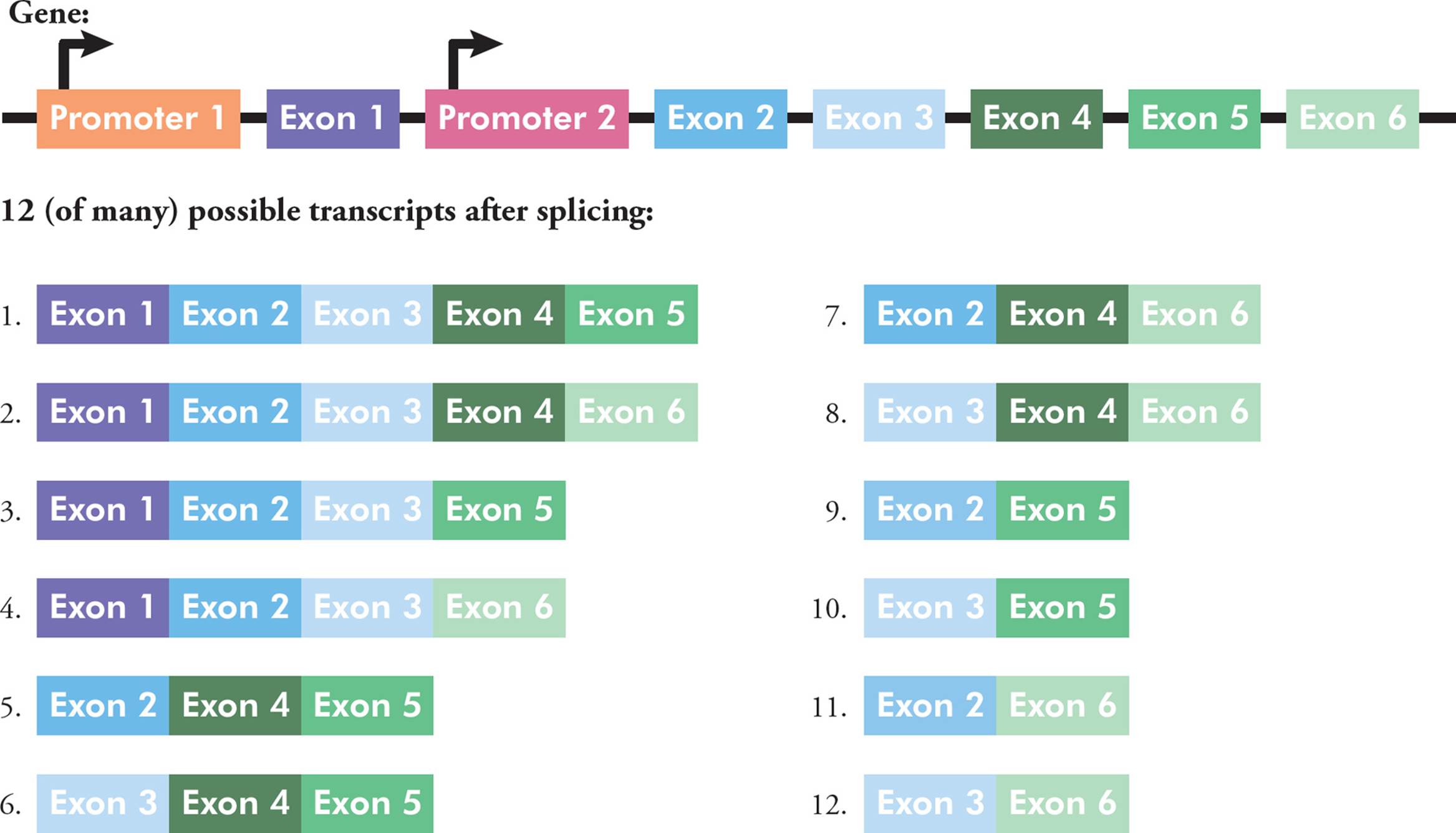
Figure 29 An Example of Alternative Splicing
Alternative splicing is mediated by introns and exons, as well as by the proteins that can bind to these sequences. There are almost 200,000 introns in the human genome, with an average of about seven per gene. It was initially thought that introns were unimportant and had no function. While it’s true that a lot of intron sequences are probably junk, the current picture of introns is a little more complicated than first believed.
Eukaryotic hnRNA must be modified in two other ways before translation can occur. A tag is added to each end of the molecule: a 5′ cap and a 3′ poly-A tail. The 5′ cap is a methylated guanine nucleotide stuck on the 5′ end [which is the end made __ (first or last?)50]. The poly-A tail is a string of several hundred adenine nucleotides. The cap is essential for translation, while both the cap and the poly-A tail are important in preventing digestion of the mRNA by exonucleases that are free in the cell.
• Why would active exonucleases be floating free in the cell?51
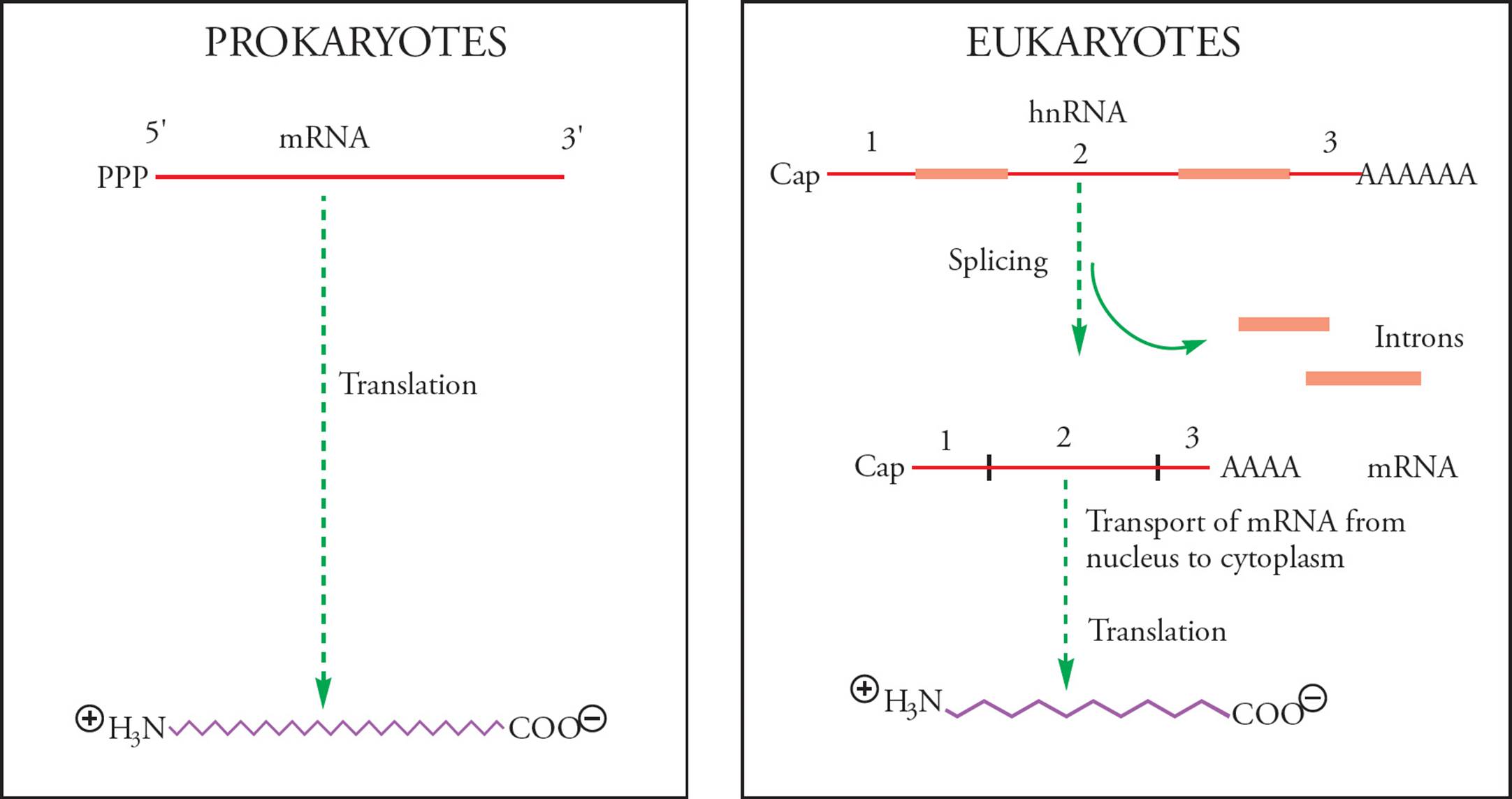
Figure 30 Comparison of Prokaryotic and Eukaryotic Gene Expression
• One piece of RNA isolated from a human cell is found to produce two different polypeptides when added to a cell-free protein synthesis system containing all the enzymes necessary for eukaryotic gene expression. When the two polypeptides are separated and digested with trypsin, they produce fragments of the following molecular weights:
Polypeptide 1: 5 kD, 8 kD, 12 kD, and 14 kD
Polypeptide 2: 3 kD, 5 kD, 8 kD, 10 kD, 12 kD, and 14 kD
How can we explain the synthesis of two different polypeptides from one piece of RNA?52
RNA Polymerase
In prokaryotes, all RNA is made by the α2ββ'σ RNA polymerase complex. In eukaryotes, there are many different RNA polymerases:
• RNA polymerase I transcribes most rRNA
• RNA polymerase II transcribes hnRNA (so ultimately mRNA), most snRNA and some miRNA
• RNA polymerase III transcribes tRNA, long ncRNA, siRNA, some miRNA and a subset of rRNA
Please note: In our discussion of replication you learned about many prokaryotic DNA polymerases. In contrast, here you learned about many eukaryotic RNA polymerases. Don’t get mixed up!
5.8 GENE EXPRESSION: TRANSLATION
Translation is the synthesis of polypeptides according to the amino acid sequence dictated by the sequence of codons in mRNA. During translation, an mRNA molecule attaches to a ribosome at a specific codon, and the appropriate amino acid is delivered by a tRNA molecule. Then the second amino acid is delivered by another tRNA. Then the ribosome binds the two amino acids together, creating a dipeptide. This process is repeated until the polypeptide is complete, at which point the ribosome drops the mRNA and the new polypeptide departs.
Transfer RNA (tRNA)
Each tRNA is composed of a single transcript produced by RNA polymerase III. The tertiary structure of every tRNA molecule is similar. tRNAs have a stem-and-loop structure stabilized by hydrogen bonds between bases on neighboring segments of the RNA chain (Figures 31 and 32). Several modified nucleotides are found in tRNA (e.g., dihydrouridine). One end of the structure is responsible for recognizing the mRNA codon to be translated. This is the anticodon, a sequence of three ribonucleotides which is complementary to the mRNA codon the tRNA translates. A key step in translation is specific base pairing between the tRNA anticodon and the mRNA codon. It is this specificity that dictates which amino acid of the twenty will be added to a growing polypeptide chain by the ribosome. [Is it likely that the three nucleotides of the anticodon contribute to the tertiary structure of tRNA by base-pairing with other nucleotides in the chain?53] The other end of the tRNA molecule has the amino acid acceptor site, which is where the amino acid is attached to the tRNA. [If you analyzed a thousand tRNA molecules, which region would you expect to vary the most?54] Since there is a tRNA for each codon, each tRNA is specific for one amino acid, while each amino acid may have several tRNAs. Each tRNA can be named according to the amino acid it’s specific for. For example, a tRNA for valine would be written tRNAVal. When the amino acid is attached, the tRNA is written this way: Val-tRNAVal.
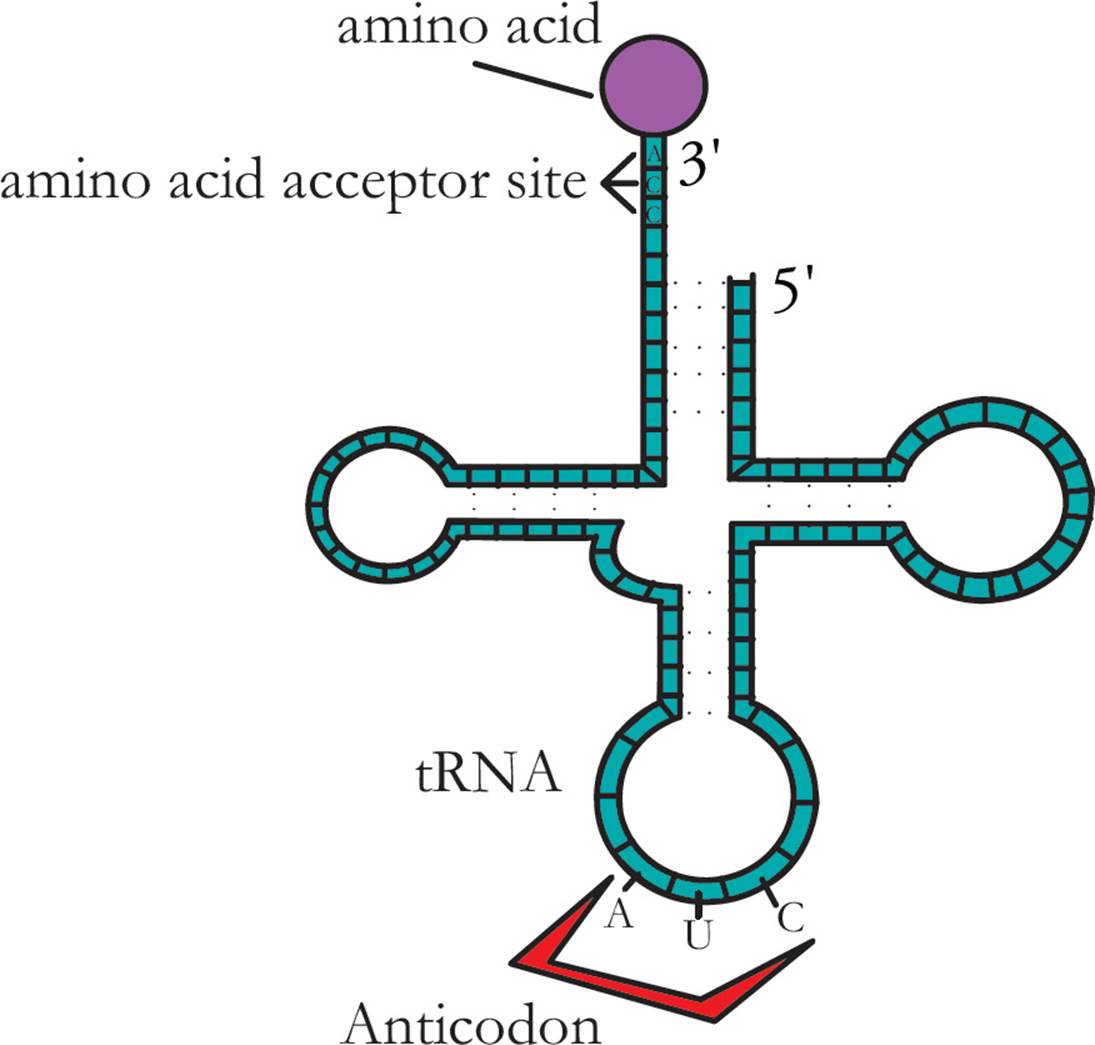
Figure 31 Cloverleaf (Two-Dimensional) Structure of tRNA
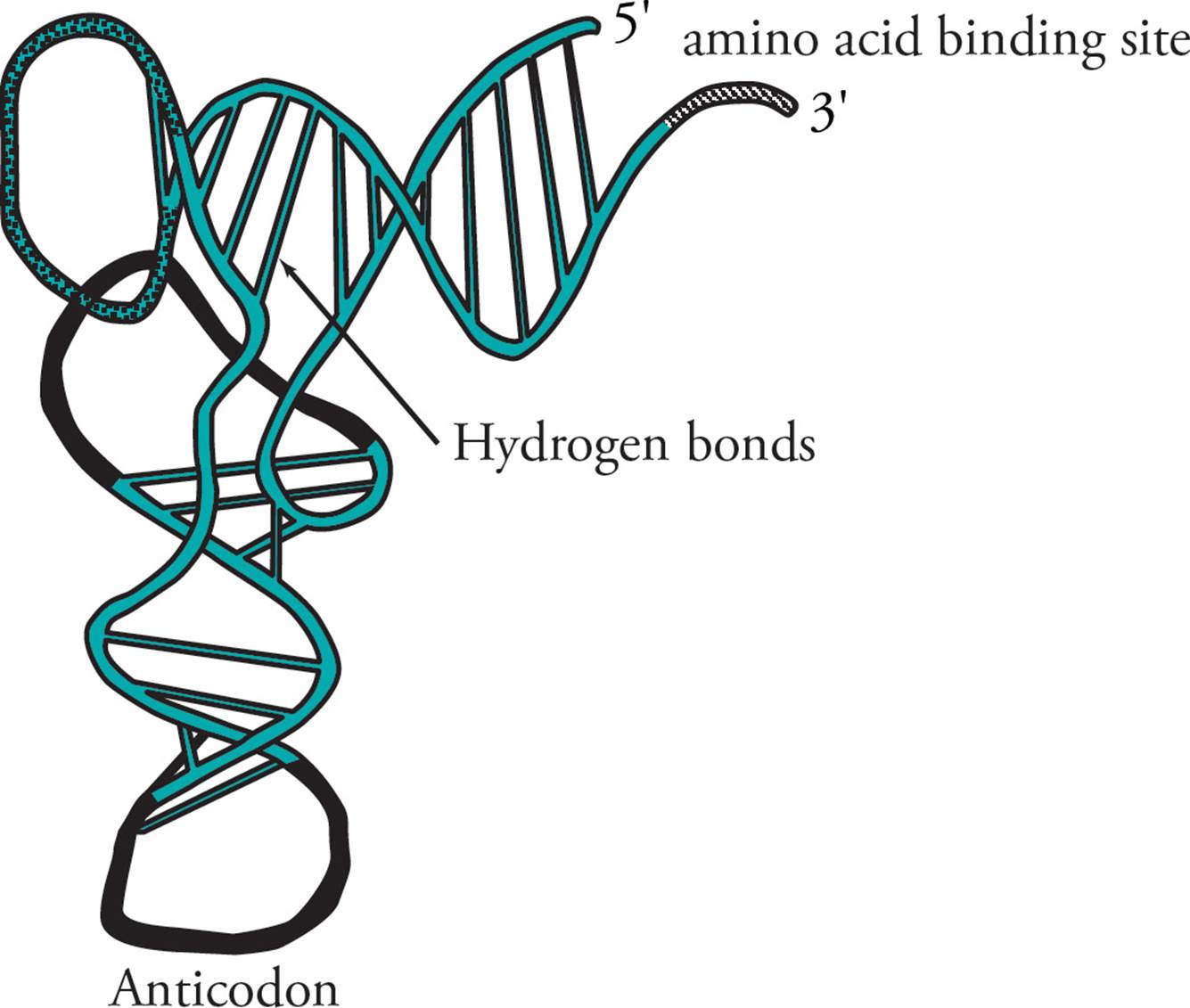
Figure 32 Three-Dimensional Structure of tRNA
tRNA molecules often contain nitrogenous bases in many positions that have been covalently modified. Base methylation is particularly common. Some specific examples are inosine (derived from adenine), pseudouridine (derived from uracil) or lysidine (derived from cytosine). Inosine in particular plays an important role in wobble base pairing.
The Wobble Hypothesis
Using the standard genetic code, you would guess that organisms have 61 distinct tRNA molecules to recognize the 61 amino acid-coding codons possible in mRNA. In actual fact, most organisms have fewer than 45 different types of tRNAs, meaning some anticodons must pair with more than one codon. Francis Crick’s Wobble Hypothesis explains this, and states that the first two codon-anticodon pairs obey normal base pairing rules, but the third position is more flexible (Figure 33). This allows for non-traditional pairing, and explains why a smaller number of tRNAs are possible.
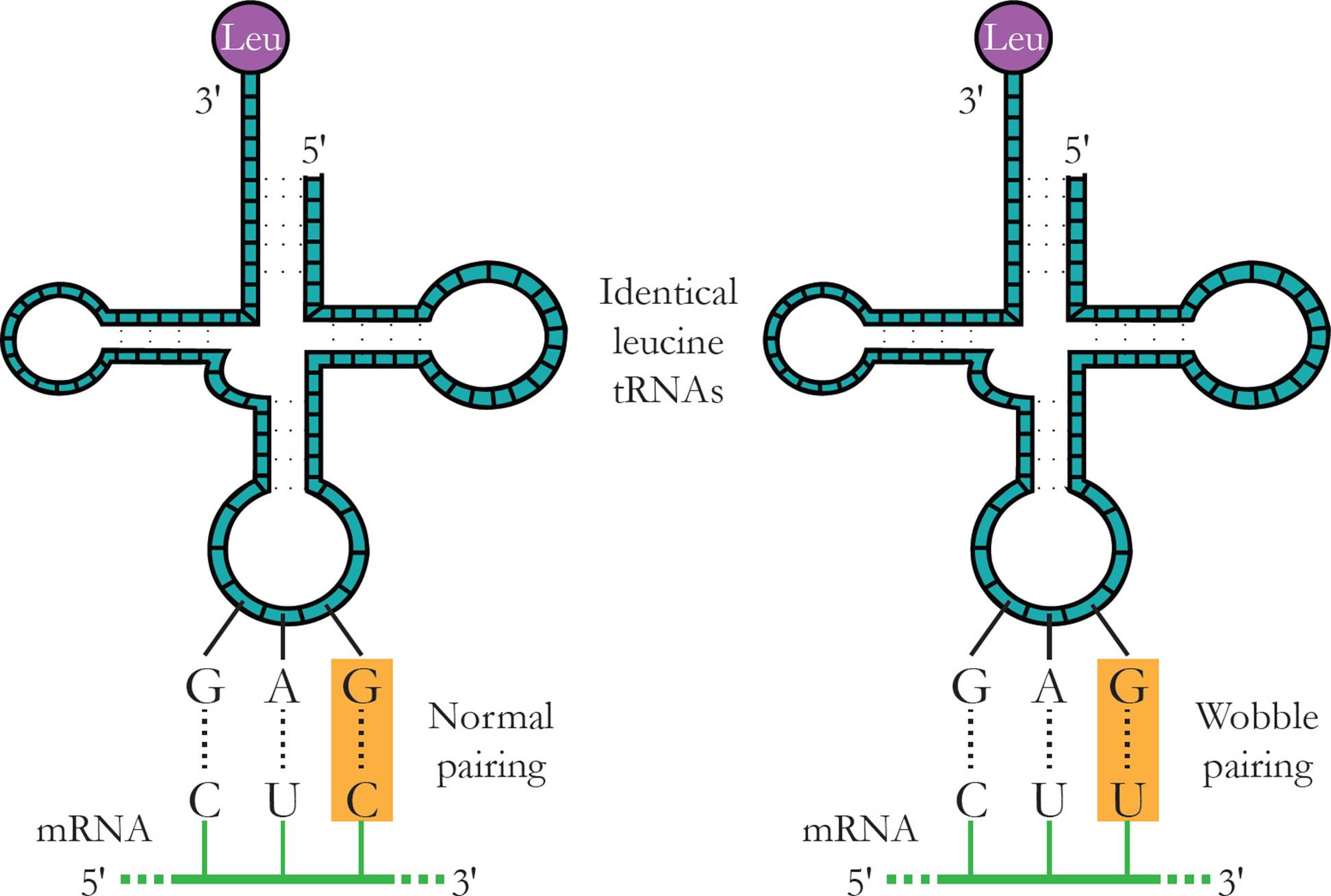
Figure 33 Wobble Base Pairing Between a tRNA Anticodon and an mRNA Codon
A modified inosine base (I) at the 5′ end of the anticodon is particularly wobbly, as it can bond to three different codon bases (A, U or C). Some common wobble pairing combinations are:
|
5′ Base in Anticodon (tRNA) |
3′ Base in Codon (mRNA) |
|
G |
C (Watson-Crick base) or U (wobble base) |
|
C |
G |
|
A |
U |
|
U |
A (Watson-Crick base) or G (wobble base) |
|
I |
A, U or C (all wobble bases) |
In other words, the most common wobble base pairs are guanine-uracil, inosine-uracil, inosine-adenine, and inosine-cytosine (G-U, I-U, I-A and I-C). Both the wobble base pair and the normal Watson-Crick base pair have similar thermodynamic stabilities.
Amino Acid Activation
Peptide bond formation during protein synthesis is a process that requires a lot of energy because the peptide bond has unfavorable thermodynamics (∆G > 0) and slow kinetics (high activation energy). Reaction coupling is used to power the process: two high-energy phosphate bonds are hydrolyzed to provide the energy to attach an amino acid to its tRNA molecule. This process is called tRNA loading or amino acid activation, and is useful because breaking the aminoacyl-tRNA bond will drive peptide bond formation forward. Amino acid activation occurs in several steps:
1) An amino acid is attached to AMP to form aminoacyl AMP. In this reaction, the nucleophile is the acidic oxygen of the amino acid, and the leaving group is PPi.
2) The pyrophosphate leaving group is hydrolyzed to 2 orthophosphates. This reaction is highly favorable (∆G << 0).
3) tRNA loading, an unfavorable reation, is driven forward by the destruction of the high- energy aminoacyl—AMP bond created in Step 1.
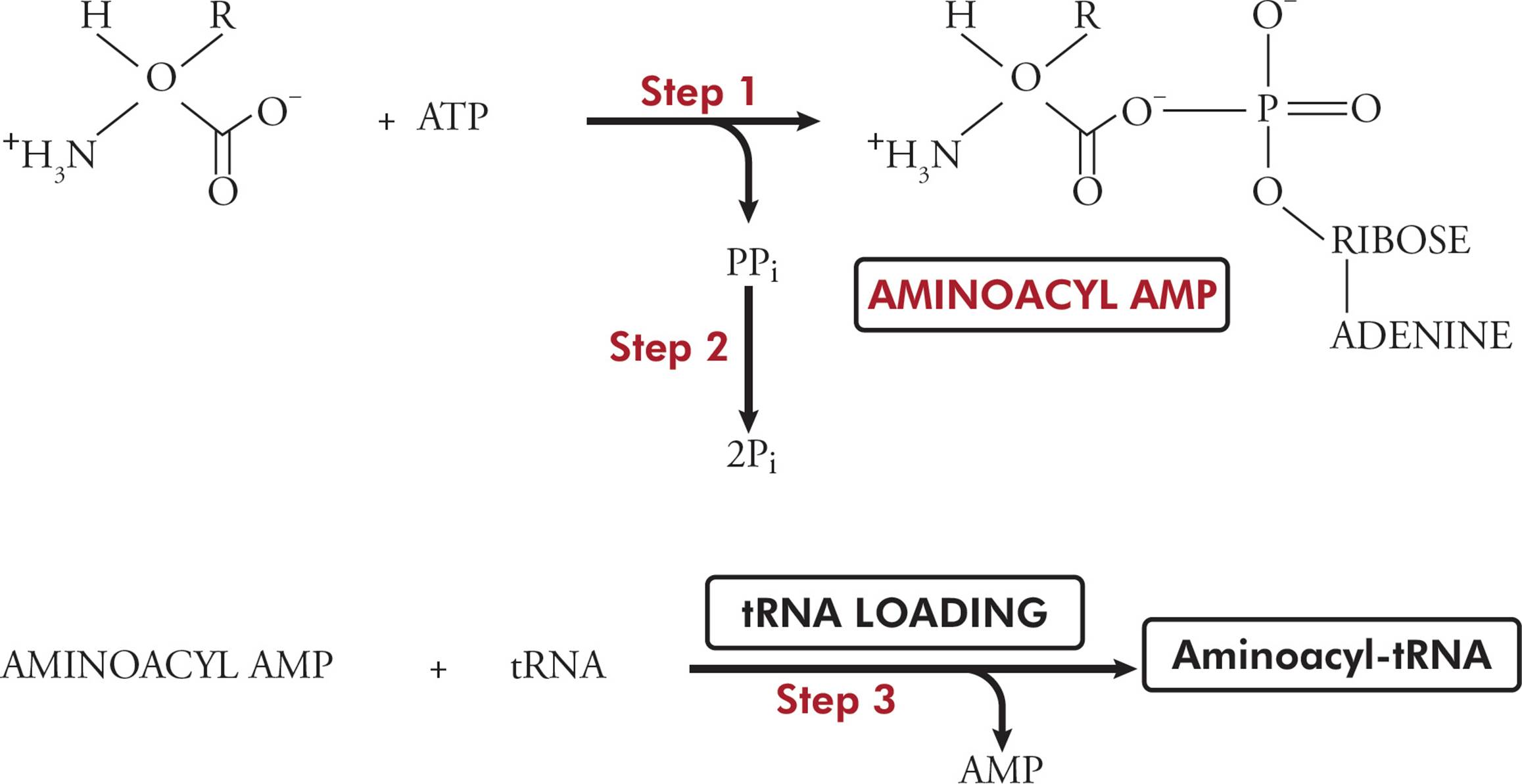
Figure 34 Amino Acid Activation as an Example of Reaction Coupling Note: water as a reactant has been left out of all reactions in this figure.
Overall, amino acid activation requires 2 ATP equivalents because it uses two high-energy bonds. An ATP equivalent is a single high-energy phosphate bond. You can get 2 ATP equivalents by hydrolyzing 2 ATP to 2 ADP + 2 Pi or by hydrolyzing 1 ATP to AMP + 2 Pi.
Eventually, the bond between the amino acid and the tRNA molecule will be broken. This hydrolysis will power peptide bond formation: the nitrogen of another amino acid will nucleophilically attack the carbonyl carbon of this amino acid, and tRNA will be the leaving group.
Aminoacyl-tRNA Synthetases
We have stated that incorporation of the appropriate amino acid in a growing polypeptide depends on the delivery of the correct amino acid by a specific tRNA. But we also noted that the amino acid acceptor sites of all tRNA molecules are the same. How is the attachment of the appropriate amino acid to each tRNA molecule accomplished? Aminoacyl-tRNA synthetase enzymes are specific to each amino acid, and there is at least one aminoacyl-tRNA synthetase for every amino acid. This family of enzymes recognizes both the tRNA and the amino acid, based on their three-dimensional structures. They are highly specific, which is important because joining the wrong amino acid to a tRNA would result in the wrong amino acid being incorporated into a polypeptide. Given that some amino acids differ only by a single methyl group, this specificity is quite amazing. Aminoacyl-tRNA synthetases also function with a very low error rate. [If there is a 1/1000 error rate in amino acid incorporation, what percentage of polypeptides that are 500 amino acid residues long will not contain any errors?55]
Overall then, amino acid activation serves two functions. One is specific and accurate amino acid delivery, and the other is thermodynamic activation of the amino acid.
• A bacterial strain with a point mutation in the gene for hexokinase is not able to metabolize glucose. The mutation causes a substitution of arginine for serine. These bacteria are used to test whether chemicals are mutagenic. The chemical is added to a culture of bacteria with glucose as the only carbon source. Any bacteria that grow must have undergone a mutation which remedied the problem (this is called suppression of the original mutation). When a particular hair spray ingredient is tested, several colonies grow on the glucose-only medium. Which one of the following might act as a suppressor of the first mutation?56
A) A point mutation during replication of a tRNA gene
B) A mutation in RNA polymerase that increases the rate of promoter recognition
C) A base pair deletion in the hexokinase gene
D) A point mutation during transcription of a tRNA molecule
The Ribosome
The ribosome is composed of many polypeptides and rRNA chains held together in a massive quaternary structure. Ribosomes float around in the cytoplasm, and each has a small subunit and a large subunit. The unit of measurement is the Svedberg, or S, unit. Svedbergs are a sedimentation rate, that is, how quickly something will sink in a gradient during centrifugation, and the units are not additive.
The prokaryotic ribosome sediments in a gradient at a rate of 70S, so it is referred to as the 70S ribosome (Figure 35). It is composed of a 30S small subunit and a 50S large subunit. The small subunit is made of a 16S rRNA and 21 peptides. Two rRNA molecules (23S and 5S) and 31 peptides make up the large subunit.
Eukaryotes have an 80S ribosome. It also has a small and large subunit. The large subunit is has three rRNA molecules (5S, 5.8S, and 28S) and 46 peptides, and sediments in a gradient at a rate of 60S. The small subunit has 33 peptides and one rRNA (18S) and sediments in a gradient at a rate of 40S.
The 23S rRNA in prokaryotes and the 28S rRNA in eukaryotes have ribozyme function. They help link amino acids during protein synthesis via peptidyl transferase activity. This contributes to peptide bond formation. Notice how the ribozymic activity of the ribosome is found in the large subunit of both prokaryotic and eukaryotic ribosomes.
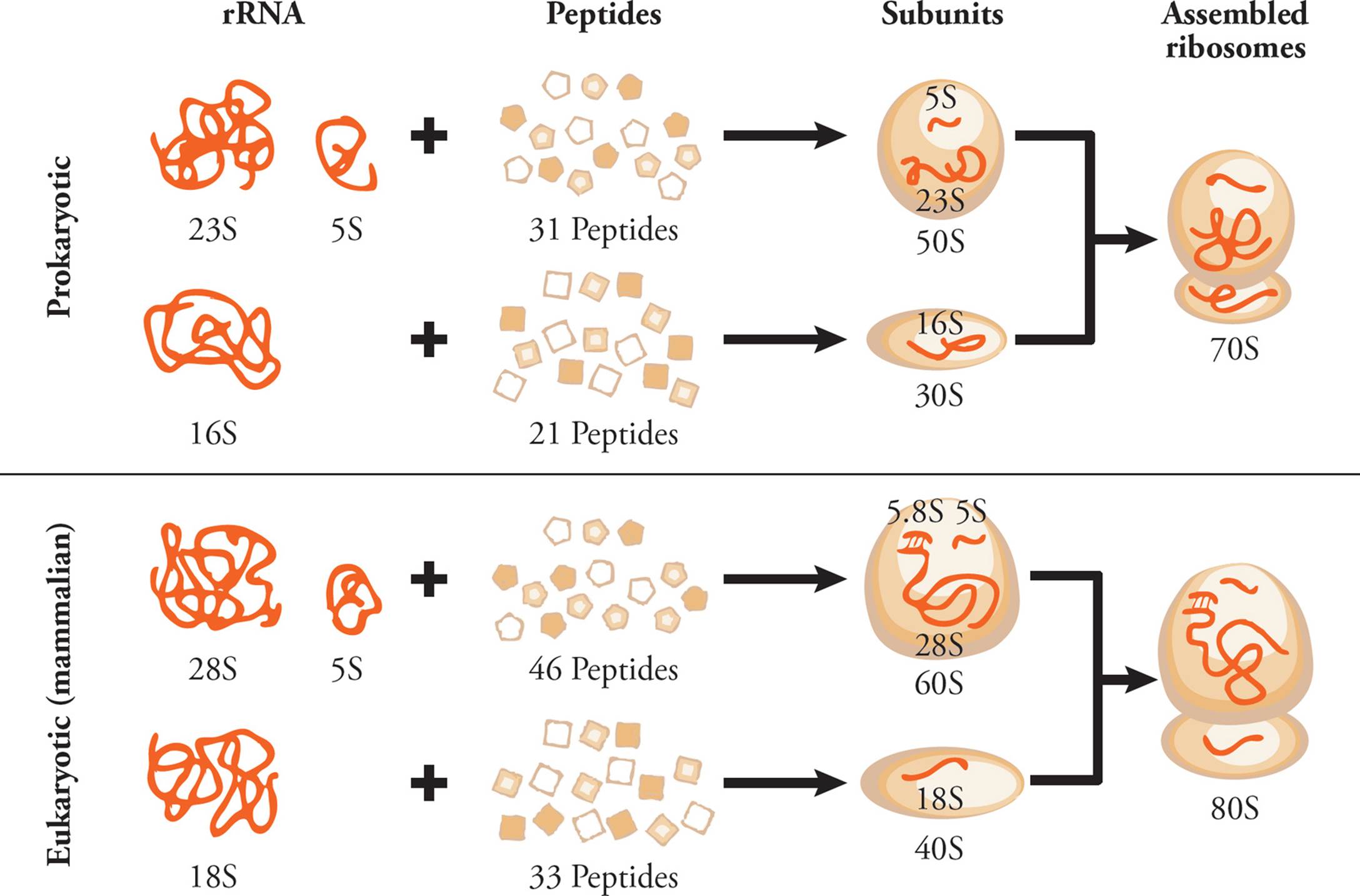
Figure 35 Ribosome Components
In both prokaryotes and eukaryotes, the complete ribosome (both subunits together) has three special binding sites. The A site (aminoacyl-tRNA site) is where each new tRNA delivers its amino acid. The P site (peptidyl-tRNA site) is where the growing polypeptide chain, still attached to a tRNA, is located during translation. The E site (exit-tRNA site) is where a now-empty tRNA sits prior to its release from the ribosome. [During translation, the next codon to be translated is exposed in the __57.] tRNAs move through the sites from A → P → E.
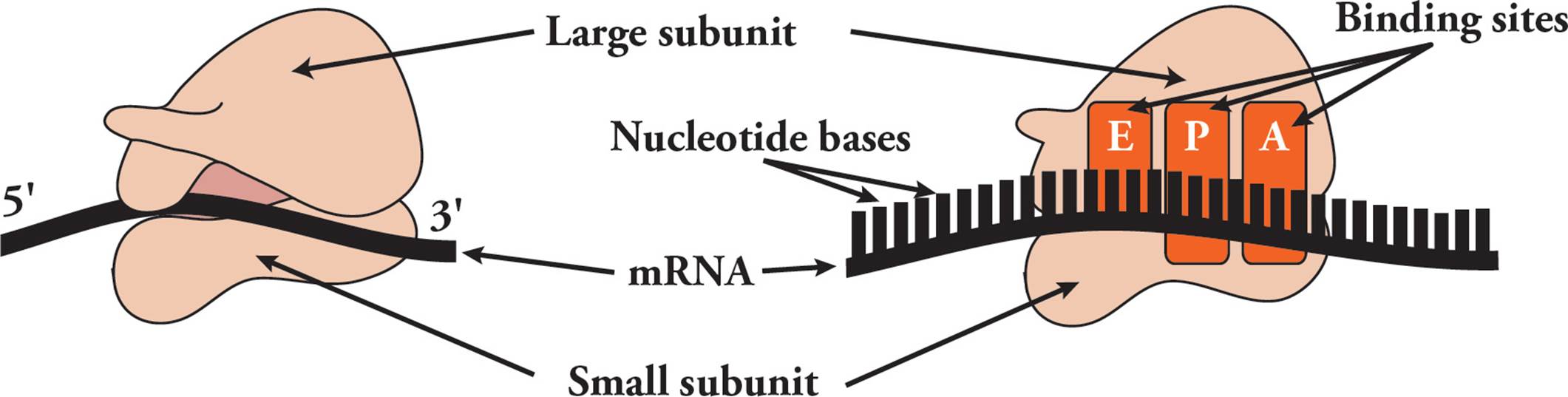
Figure 36 The Ribosome
Prokaryotic Translation
In prokaryotes, translation occurs in the same compartment and at the same time as transcription. In other words, while the mRNA is being made ribosomes attach and begin translating it. [That means that the first end of the mRNA to be translated is 5′ or 3′?58] Note that it says ribosomes above. Several ribosomes attach to the mRNA and translate it simultaneously (see Figure 37; you may hear the term polyribosome used to describe this arrangement; polyribosomes are seen in both prokaryotes and eukaryotes). [You figured out the direction of translation on the mRNA from what you already know. Do you have any previous knowledge that would help you answer this: Does translation always begin at the 5′ end of the mRNA, or somewhere up the chain?59]
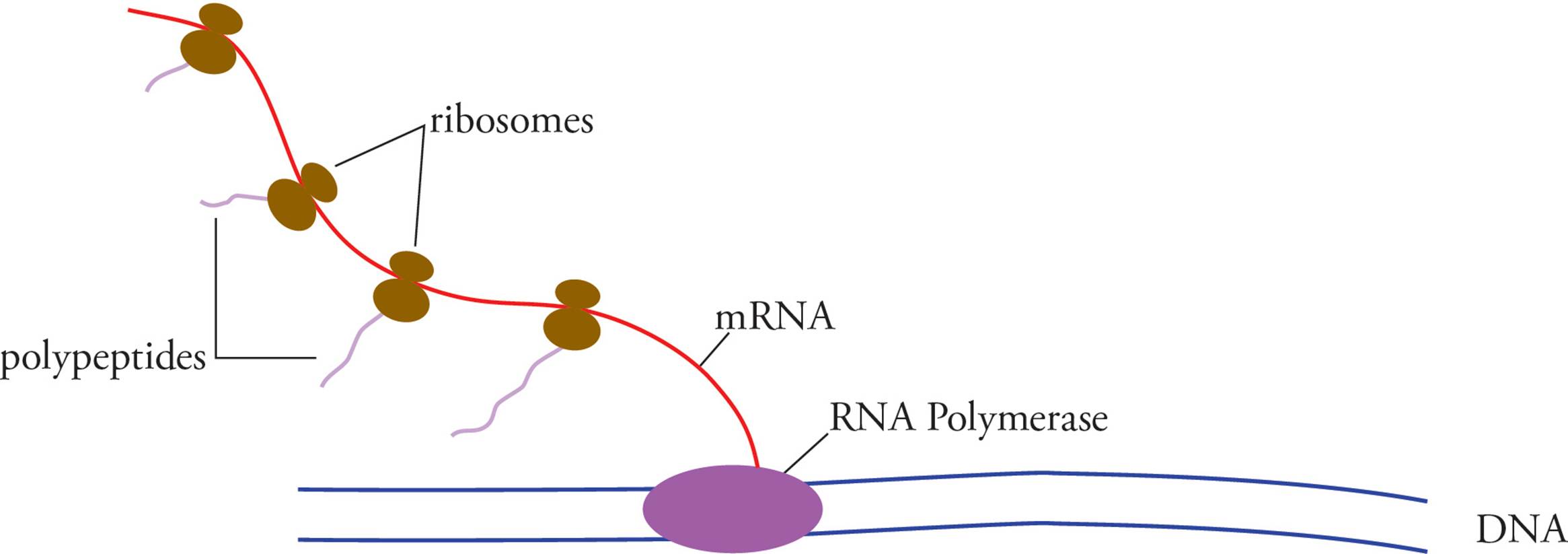
Figure 37 A Prokaryotic Polyribosome
Because prokaryotes often have polycistronic mRNAs, their ribosomes can also start translation in the middle of the chain. This means termination and initiation sequences are found between each ORF. Even for the first open reading frame on a transcript, translation doesn’t begin right at the 5′ end. An upstream regulatory sequence is essential for initiation, just as in transcription. Here, instead of a promoter, we have a ribosome binding site, also known as the Shine-Dalgarno sequence, located at –10 (ten ribonucleotides upstream, or on the 5′ side of the start codon). The Shine-Dalgarno sequence is complementary to a pyrimidine rich region on the small subunit, and thus helps position the initiation machinery on the transcript.
Like transcription, translation has three distinct stages: initiation, elongation, and termination. Many antibiotics function by inhibiting a particular stage60.
Initiation starts with the small ribosomal subunit (30S) binding two initiation proteins called IF1 and IF3. This complex then binds the mRNA transcript. Next, the first aminoacyl-tRNA joins, along with a third initiation factor called IF2, which is also bound to one GTP. Finally, the 50S subunit completes the complex. This process is powered by the hydrolysis of one GTP molecule.61 The first aminoacyl-tRNA is special; it is called the initiator tRNA, abbreviated fMet-tRNAfMet. The “fMet” stands for formylmethionine, which is a modified methionine used as the first amino acid in all prokaryotic proteins.62 The initiator tRNA sits in the P site of the 70S ribosome, hydrogen-bonded with the start codon. [What is the start codon? Does this codon initiate translation wherever it appears?63] Before elongation, all initiation factors dissociate from the complex.
Elongation, a three-step cycle, may now begin. In the first step, the second aminoacyl-tRNA enters the A site and hydrogen bonds with the second codon. This process requires the hydrolysis of one phosphate from GTP. This is done by an elongation factor protein called Tu (EF-Tu), which is a GTPase. A second elongation factor called EF-Ts removes the remaining GDP from EF-Tu, thus helping it reset. In the second step, the peptidyl transferaseactivity of the large ribosomal subunit (the 23S rRNA) catalyzes the formation of a peptide bond between fMet and the second amino acid. The amino group of amino acid #2 acts as nucleophile, and tRNAfMet is the leaving group; it dissociates from the ribosome. A new dipeptide is now attached to tRNA #2. Now you can Figure out the direction of translation from the point of view of the polypeptide; you won’t have to memorize it.64 The third step is translocation, in which tRNA #1 (now empty) moves into the E site, tRNA #2 (holding the growing peptide) moves into the P site, and the next codon to be translated moves into the A site. Elongation factor G (EF-G) helps with translocation, and this process costs one GTP. EF-G is sometimes called a translocase because of its function in this step. The new dipeptide is still attached to tRNA #2, and tRNA #2 is still H-bonded to codon #2. The presence of tRNA #1 in the E site (still H-bonded to codon #1), is thought to help maintain the reading frame of the mRNA (disruption of tRNA binding to the E site results in an increase in the number of frameshift mutations in the resulting protein). EF-Tu eventually helps remove this tRNA from the E site. [Does the ribosome move relative to the mRNA during translocation?65] These three steps repeat over and over again, connecting amino acids in the order their codons appear along the mRNA strand (and thus appear in the A site).
Termination occurs when a stop codon appears in the A site. Instead of a tRNA, a release factor now enters the A site. This causes the peptidyl transferase to hydrolyze the bond between the last tRNA and the completed polypeptide. Prokaryotes have three release factor proteins, which mediate translation termination by recognizing stop codons. RF1 recognizes termination codons UAA and UAG, and RF2 recognizes UAA and UGA. RF3 is a GTP-binding protein that doesn’t recognize a stop codon, but instead leads to the dissociation of RF1/RF2 after peptide release. Finally, the ribosome separates into its subunits and releases both mRNA and polypeptide.
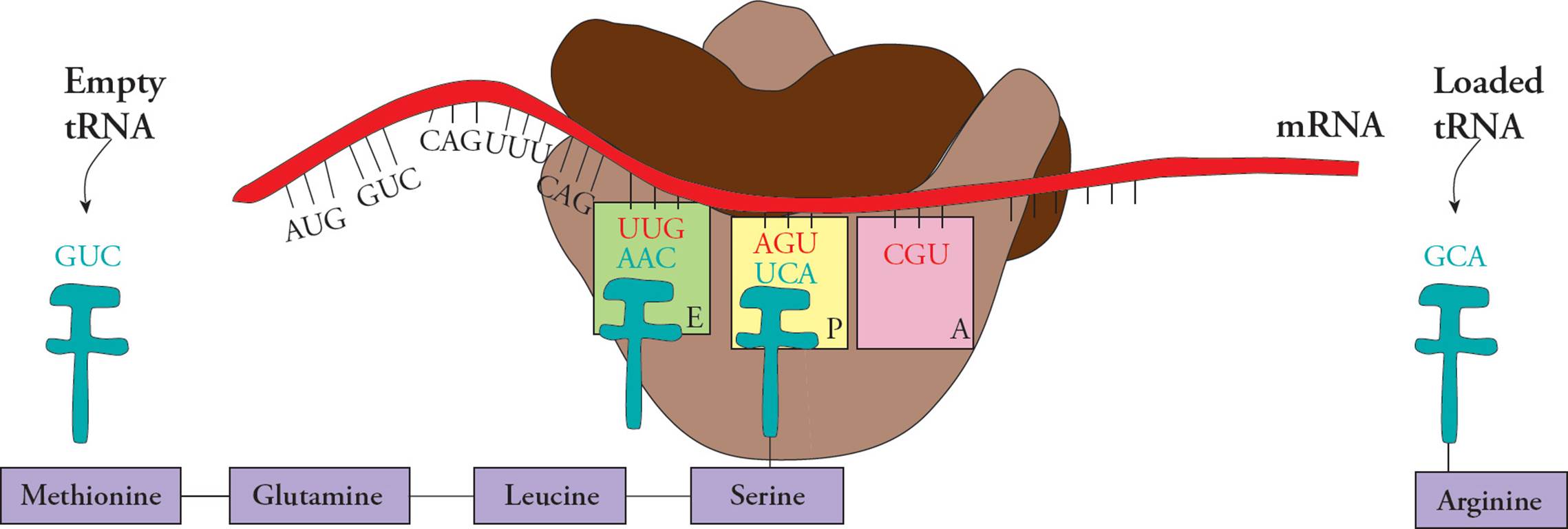
Figure 38 Translation Elongation
Let’s focus for a moment on the energetics of translation. Why doesn’t peptide bond formation require GTP hydrolysis, like the other steps in translation?66 You should be able to answer questions like this: How many high energy phosphate bonds are required to make a 50 amino acid polypeptide chain, including the energy used to activate amino acids to aminoacyl-tRNAs?67
Eukaryotic Translation
There are several differences between eukaryotic and prokaryotic translation. Many of these have already been mentioned: the ribosome is larger (80S) and has different components than the prokaryotic ribosome, the mRNA must be processed before it can be translated (spliced, with cap and tail added), and the N-terminal amino acid is different (Met instead of fMet). Also remember that eukaryotic mRNA must not only be spliced, capped, and tailed, but it also requires transport from nucleus to cytoplasm, thus transcription and translation cannot proceed simultaneously.
Eukaryotes do not use the Shine-Dalgarno sequence to initiate translation. There are 5′ UTR sequences in eukaryotes that function in starting translation; a common one is the Kozak sequence, which is a consensus sequence typically located a few nucleotides before the start codon.
Eukaryotic translation begins with formation of the initiation complex. First, a 43S pre-initiation complex forms, composed of the 40S small ribosomal submit, Met-tRNAMet, and several proteins called eukaryotic initiation factors (or eIFs). Next, this assembled complex is recruited to the 5′ capped end of the transcript, by an initiation complex of proteins (including other eIF proteins). Additional proteins are recruited (such as a polyA tail binding protein) and the initiation complex starts scanning the mRNA from the 5′ end, looking for a start codon. Once the start codon has been found, the large ribosomal subunit (60S) is recruited and translation can begin.
Some eIF proteins are essential to initiate translation and others help regulate the process. For example, eIF3 binds the small ribosomal subunit and prevents it from prematurely associating with the 60S subunit. The amount of eIF proteins in the cell is closely controlled, and this affects the amount of translation occurring. eIF4A is a helicase and unwinds mRNA, eIF4E binds the 5′ cap of the mRNA and eIF4G is a scaffold protein. Each of these three function in the initiation complex, and their levels are a rate-limiting step for translation. Higher amounts of these three proteins means the cell can perform more translation, while a lower amount decreases translation. Activity of eIF proteins is controlled by post-translational modification, such as phosphorylation. This couples translation to upstream cell signaling pathways.
Eukaryotes have two elongation factors. eEF-1 has two subunits, one that helps with entry of an aminoacyl tRNA into the A site and one that is a guanine nucleotide exchange factor, catalyzing the release of GDP. The eukaryotic translocase is called eEF-2. Additional elongation factors are required to facilitate peptide bond formation.
The order in which the initiation complex is formed is different in eukaryotes. [Are the nascent (newly formed) polypeptide chains emerging from a polyribosome in a eukaryote all the same?68] Eukaryotic translation termination involves two release factors. eRF1 recognizes all three termination codons, and eRF3 is a ribosome-dependent GTPase that helps eRF1 release the completed polypeptide.
• Which one of the following pairs of processes may occur simultaneously on the same RNA molecule in a eukaryotic cell?
A) Translation and transcription
B) Transcription and splicing
C) Splicing and translation
D) Messenger RNA degradation and transcription69
Cap-Independent Translation
It was long thought that all eukaryotic translation started at the 5′ end of an mRNA. In other words, all eukaryotic transcripts were assumed to be monocistronic, and coded for only one polypeptide chain. It is true that this mechanism is by far the major one in eukaryotic cells. Because of the important role of 5′ mRNA cap recognition, it’s called cap-dependent translation.
However, it’s recently been discovered that eukaryotes are sometimes capable of starting translation in the middle of an mRNA molecule, a process called cap-independent translation (because the beginning of translation doesn’t require the 5′ cap of the mRNA). To do this, the transcript must have an internal ribosome entry site, or IRES. This is a specialized nucleotide sequence, and was first discovered in viruses. Since then, IRESs have been found in a number of eukaryotic transcripts. Most code for proteins that help the cell deal with stress, or help activate apoptosis. In other words, the IRESs found so far make sure the cell can make essential proteins when under sub-optimal growth conditions. Cells under stress generally inhibit translation (via inhibiting translation initiation), and cap-independent translation allows the cell to make proteins when doing so is crucial for survival or programmed cell death. Activation of translation using an IRES requires different proteins than normal initiation.
Additional nucleotide sequences have been identified, which allow cap-independent translation in eukaryotes. While some of these are used in molecular biology labs, it’s unclear how or if they function in normal eukaryotic cells.
5.9 CONTROLLING GENE EXPRESSION
Adult humans have over 220 different types of cells, all with the same genome, but with different attributes such as morphology, lifespan, function, ability to secrete, response to signaling molecules, mobility, etc. These changes are due to differences in gene expression and protein function. In each cell type, some genes are expressed and others are silenced, further, genes that are expressed can have different levels of expression, where in one cell type the gene is expressed at a high level (to produce lots of ncRNA or protein), and in a different cell type the same gene is expressed at a low level. They can also have varying activity, stability and half-life. These variations in gene expression can be altered using many different mechanisms:
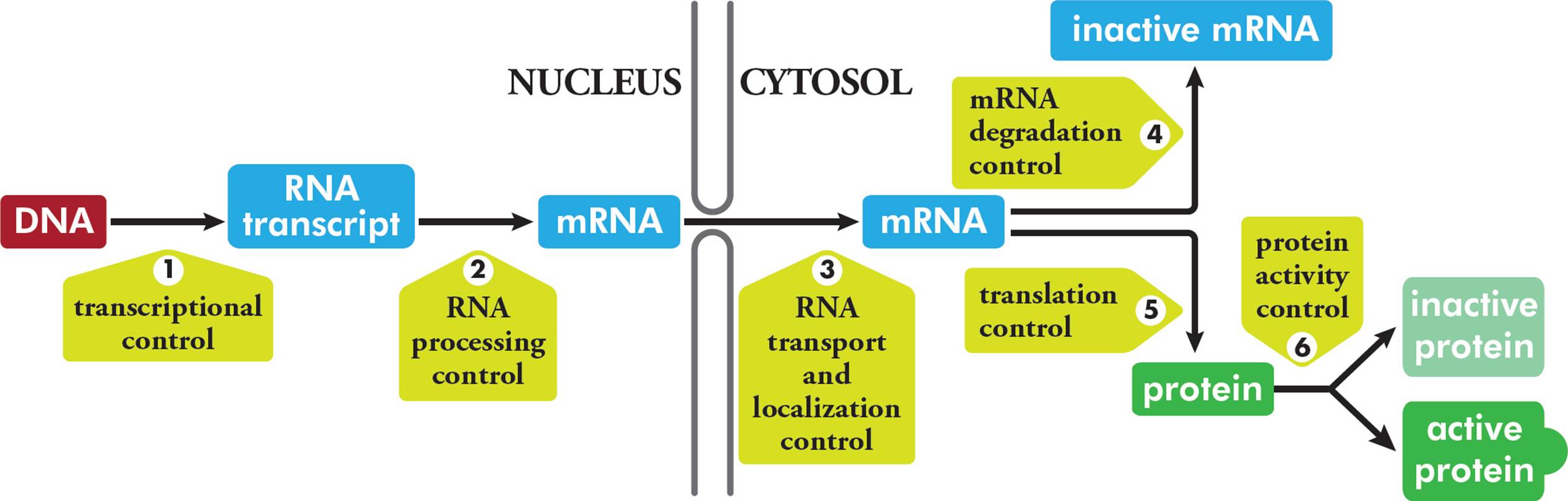
Figure 39 Mechanisms of Controlling Gene Expression in Eukaryotes
Transcription is the principle site of the regulation of gene expression in both eukaryotes and prokaryotes. This means that the amount of each protein made in every cell is affected by the amount of mRNA that gets transcribed. Gene expression can also be controlled epigenetically. Broadly speaking, epigenetics focuses on changes in gene expression that are not due to changes in DNA sequences, but are either heriTable or have a long term effect. The three most commonly studied areas in this field are DNA methylation, chromatin remodeling, and RNA interference.
DNA Methylation and Chromatin Remodeling
Both prokaryotic and eukaryotic DNA can be covalently modified by adding a methyl group. Bacteria methylate new DNA shortly after synthesis, and the brief delay is useful in mismatch repair pathways (see above). Methylation can also control gene expression in prokaryotes, either by promoting or inhibiting transcription.
Eukaryotic DNA methylation has been found in every vertebrate genome studied so far. Broadly speaking, it plays an important role in controlling gene expression (especially during embryonic development), and has also been implicated in several diseases. DNA methylation turns off eukaryotic gene expression two ways:
1) Methylation physically blocks the gene from transcriptional proteins.
2) Certain proteins bind methylated CpG groups and recruit chromatin remodeling proteins that change the winding of DNA around histones.
• Regulation of a gene is examined in vitro in the presence and absence of chromatin assembly, and in the presence and absence of a sequence-specific regulator of transcription. Transcription is quantitated after the experiment and the following results are obtained:
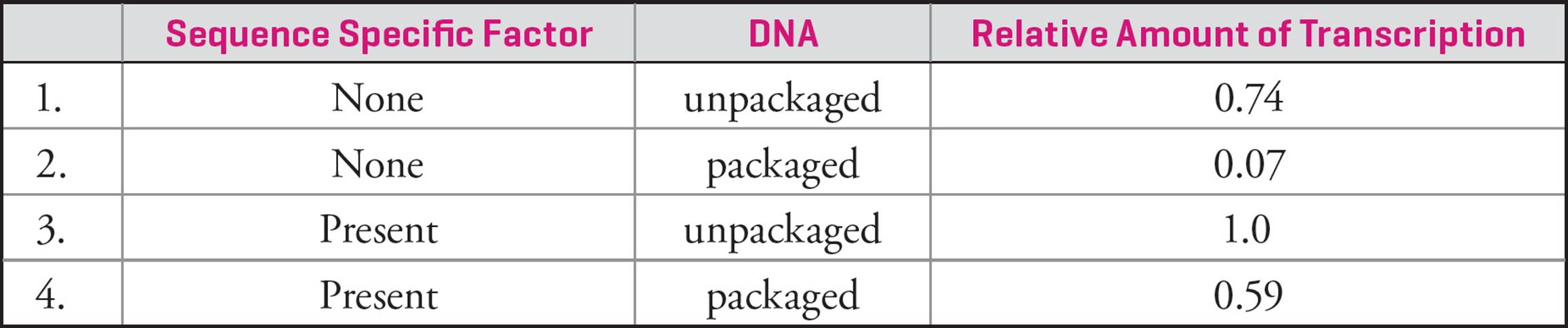
Which one of the following conclusions can be drawn from this experiment?70
A) The degree of activation by the sequence-specific factor is greater in the presence of chromatin assembly than in its absence.
B) The sequence-specific factor acts to repress transcription.
C) The histones increase the rate of transcription.
D) The sequence-specific factor increases the rate of transition from a closed complex to an open complex.
Gene Dose
One way to increase gene expression is to increase the copy number of a gene by amplification. Increasing gene dose will allow a cell to make large quantities of the corresponding protein. Similarly, gene deletion causes a decrease in gene expression. Both are examples of copy number variation, discussed in Section 5.2.
Variations on Diploid Gene Expression
Since we are diploid organisms, we have two copies of every gene. In most cases, both are either expressed or not. (Genes that are not expressed in a certain tissue or cell are said to be silenced.) However, there are some exceptions to this.
Imprinting
Genomic imprinting is when only one allele of a gene is expressed. In some situations, the maternal allele is expressed, and in others the paternal allele is expressed. Imprinted genes tend to be clustered together on chromosomes. Imprinting is a dynamic process and can change from generation to generation. In other words, a gene that is imprinted in an adult may be “unimprinted” and expressed in that adult’s offspring. This observation led to the notion that imprinting is an epigenetic process. Silencing of a certain gene involves DNA methylation, histone modification, and binding of long ncRNAs. These epigenetic marks are established in the germline, and are maintained throughout life and mitotic divisions.
X Chromosome Inactivation
Female mammals have two X chromosomes, one of which is active (called Xa) and one of which is silenced, or inactive (and is called Xi). In humans, X-inactivation occurs early in development, at the blastocyst stage (Chapter 14). Each cell in the inner cell mass randomly inactivates an X chromosome, and this decision is irreversible. This means every cell derived from each cell in the inner cell mass will have the same X chromosome inactivated, however, because each cell makes its own decision, an adult can have different X chromosomes inactivated in different tissues and cells. Because of X-inactivation, all humans have the same number of gene products for the X chromosome; males have only one X chromosome, and females have only one active X chromosome. Not all animals behave the same when it comes to X-inactivation. Some animals (such as marsupials) consistently silence one X chromosome; in the case of marsupials, the paternally derived X chromosome is inactivated and the maternal X chromosome is active. Xi is very condensed, and packaged in heterochromatin. It has high levels of DNA methylation.
Regulation of Transcription: Prokaryotes
Regulation of transcription is the primary method of regulation of gene expression in prokaryotes. One simple mechanism of transcriptional regulation in bacteria is that some promoters are simply stronger than others. The problem with this mechanism of regulation is that it is “pre-set” and cannot respond to changing conditions within the cell. Bacteria also possess far more complex regulatory mechanisms, which activate or suppress transcription depending on current needs for specific gene products. For example, bacteria only produce the enzyme β-galactosidase and other proteins required for lactose catabolism when lactose is present. [Assuming these protein products do not have a harmful effect on the cell, what advantage might there be in turning off the genes when the protein products are not required?71]
• Are the terms polypeptide enzyme and gene product synonymous? Or are there gene products that are not polypeptide enzymes? Are there polypeptides which are not enzymes?72
Enzymes involved in anabolism (biosynthesis) should be produced when the item they help make (their product) is scarce. Enzymes involved in catabolism (degradative metabolism) should be produced when the item they help break down (their substrate) is abundant, such as food. Hence there are two basic ways we can imagine how transcription is regulated. The transcription of enzymes involved in biosynthetic pathways should be inhibited by their product. The transcription of enzymes involved in catabolic pathways should be automatically inhibited whenever the substrate is not around, and activated when it is. That is in fact exactly what happens. Anabolic enzymes whose transcription is inhibited in the presence of excess amounts of product are repressible. Catabolic enzymes whose transcription can be stimulated by the abundance of a substrate are called inducible enzymes.73
There are two common examples of this. The lac operon is inducible, since the enzymes it codes for are part of lactose catabolism, and the trp operon is repressible, since the enzymes it codes for mediate tryptophan biosynthesis or anabolism. An operon has two components, a coding sequence for enzymes, and upstream regulatory sequences or control sites. Operons may also include genes for regulatory proteins, such as repressors or activators, but don’t have to. These genes can be located elsewhere in the genome and typically have their own promoters.
The Lac Operon
The lac operon contains several components:
1) P region: the promoter site on DNA to which RNA polymerase binds to initiate transcription of Y, Z, and A genes
2) O region: the operator site to which the Lac repressor binds
3) Z gene: codes for the enzyme β-galactosidase, which cleaves lactose into glucose and galactose
4) Y gene: codes for permease, a protein which transports lactose into the cell
5) A gene: codes for transacetylase, an enzyme which transfers an acetyl group from acetyl-CoA to β-galactosides (note that this function is not required for lactose metabolism)
Additionally, there are two genes, each with their own promoter, that code for proteins important in the regulation of the lac operon:
1) crp gene: located at a distant site, this gene codes for a catabolite activator protein (CAP) and helps couple the lac operon to glucose levels in the cell
2) I gene: located at a distant site, this gene codes for the Lac repressor protein
So overall, there are five protein coding genes and two regulatory sequences. Both crp and I have their own promoters. The protein products of these two genes control gene expression of Z, Y and A.
Bacterial cells preferentially use glucose as an energy source. This means that in the presence of glucose, the lac operon will be off, or expressed at low amounts (see Figures 40 and 41). This is mediated by the CAP and repressor proteins. Glucose levels control a protein called adenylyl cyclase, which converts ATP to cAMP. In high glucose conditions, adenylyl cyclase is inactivated and cAMP levels are very low. In low glucose conditions, the opposite is true: adenylyl cyclase is activated and cAMP levels are high. CAP binds cAMP and this complex binds the promoter of the lac operon (Figure 42). This helps activate RNA polymerase at the lac operon and contributes to the operon being turned on when glucose levels are low.
The I gene codes for a repressor protein, which binds the operator of the lac operon. This prevents RNA pol from binding the promoter and transcribing Z, Y, and A genes, thereby blocking transcription of the operon when lactose is absent (Figure 40). The repressor protein can also bind lactose, and this blocks its activity on the operator. This binding is allosteric, meaning it happens at a distant site from operator binding. It causes a conformational change in the tertiary structure of the repressor protein, such that it is no longer capable of binding to the operator. As a consequence, it falls off the DNA (Figures 41 and 42).
High transcription of Z, Y, and A genes occurs when glucose is absent and lactose is present (Figure 42). Low glucose results in an increased amount of cAMP, which binds to CAP and helps activate RNA polymerase activity at the lac operon. Lactose presence means the Lac repressor protein is unable to bind the lac operator and negatively regulate transcription, thus the polycistronic mRNA is transcribed at high levels. When the supply of lactose becomes very scarce, there isn’t enough to bind to the repressors, and most of the repressor proteins return to their original structure. They now rebind to the operator, decreasing transcription of Z, Y, and A genes.
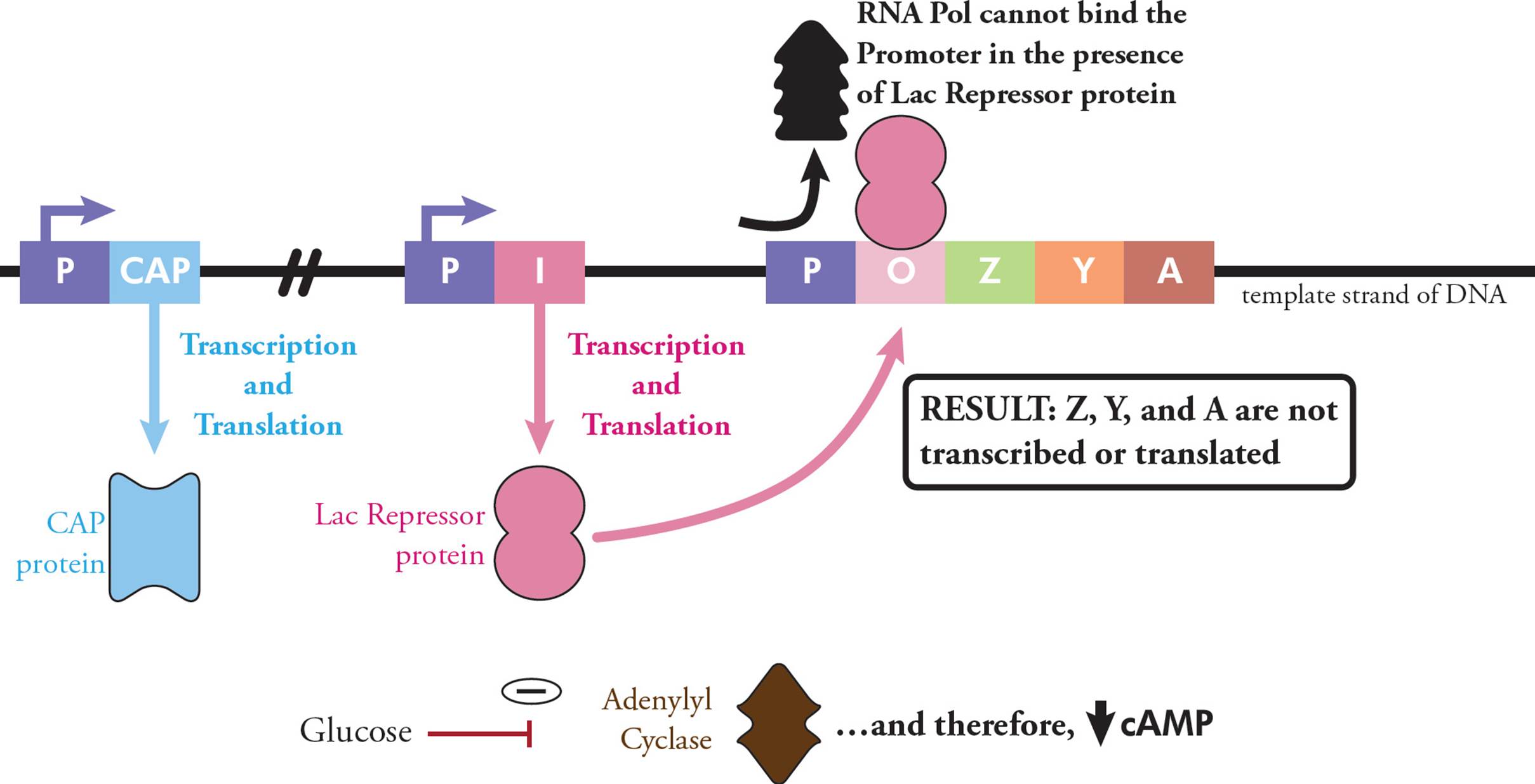
Figure 40 The Lac Operon in the Presence of Glucose and Absence of Lactose
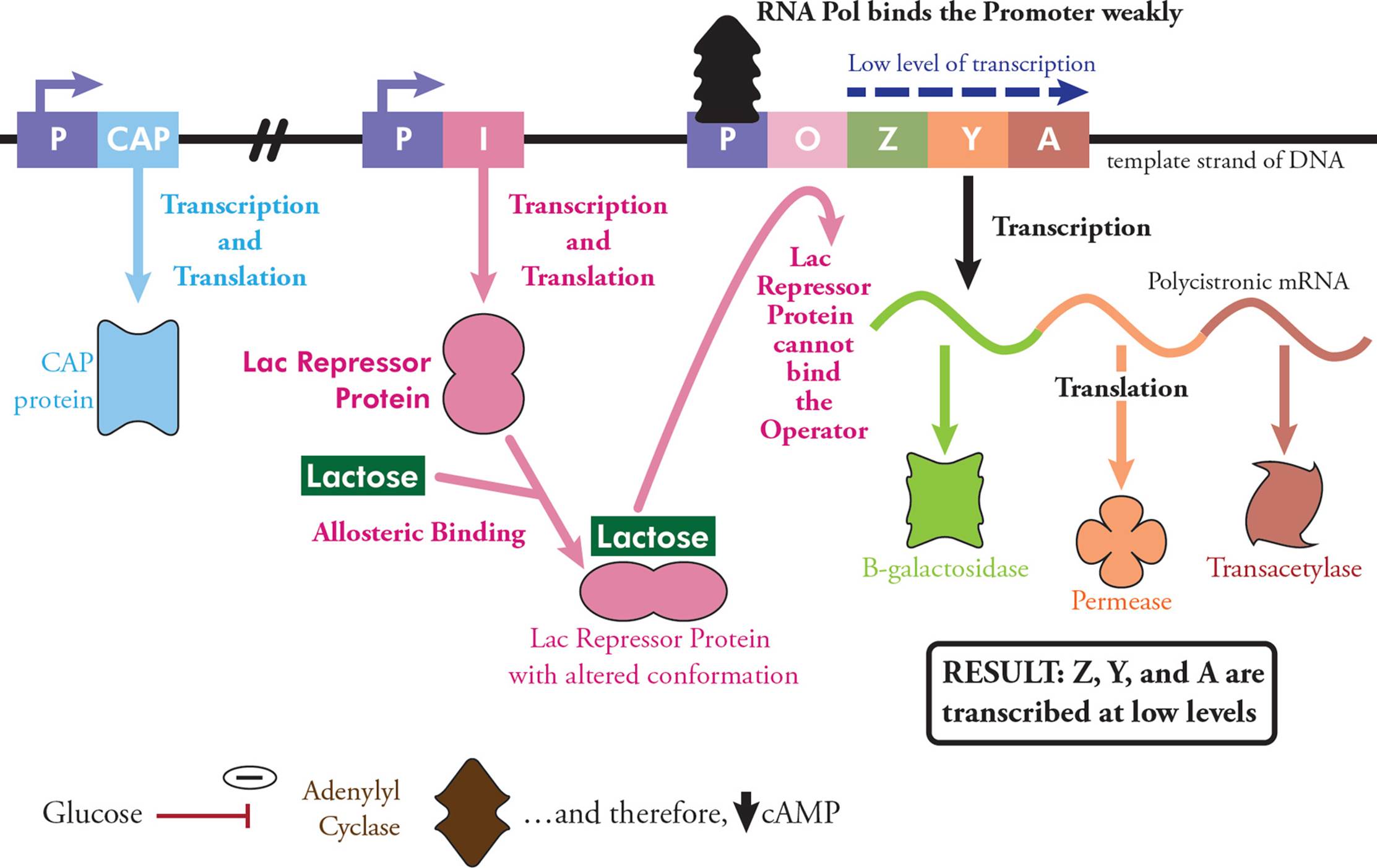
Figure 41 The Lac Operon in the Presence of both Glucose and Lactose
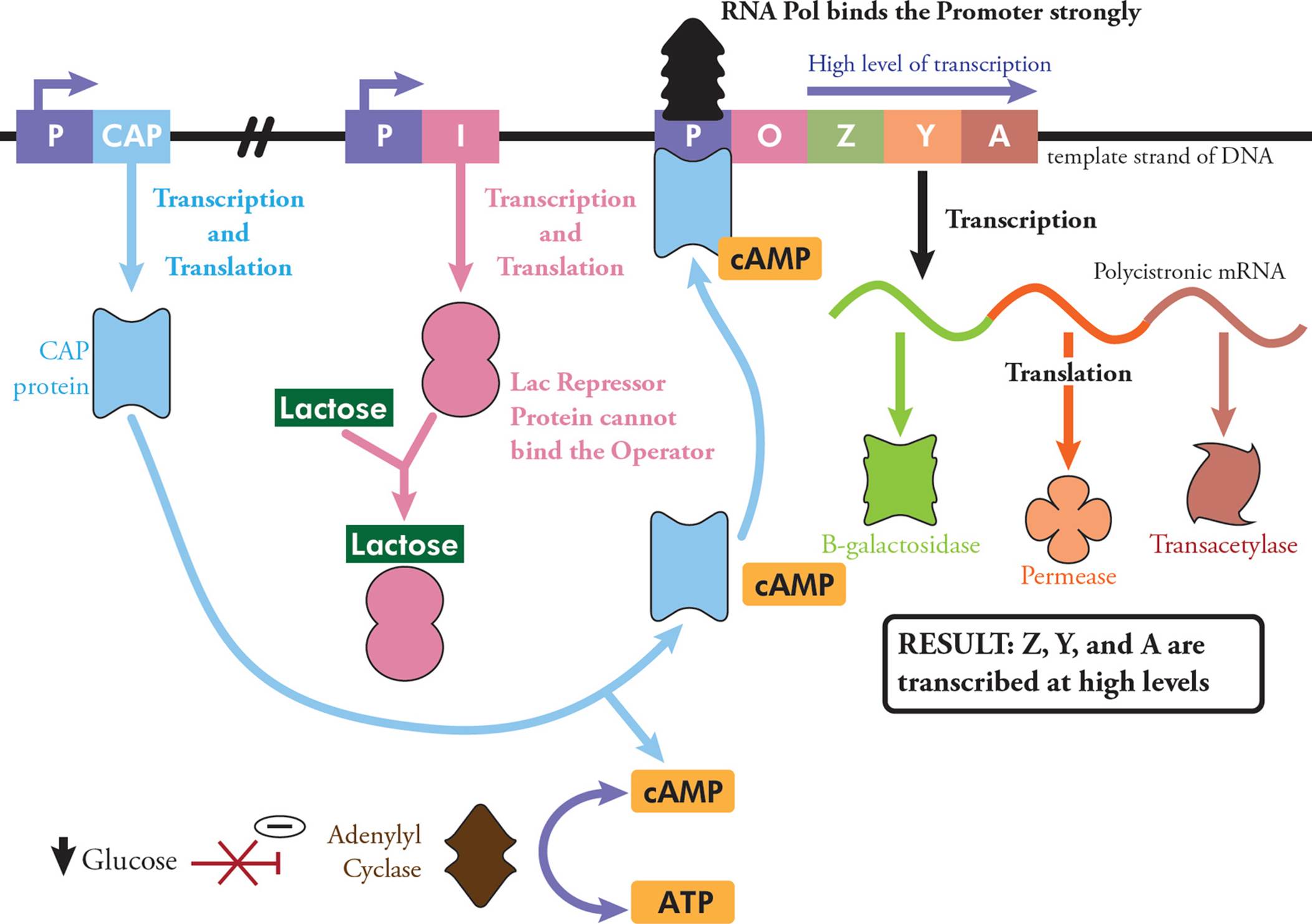
Figure 42 The Lac Operon in the Absence of Glucose and Presence of Lactose
• If the operator is mutated so that the lac repressor can no longer bind, what effect will this have on transcription?74
A) Transcription of Gene Z will be activated, and Genes Y and A will not be affected.
B) None of the genes will be transcribed, regardless of the presence or absence of lac repressor.
C) Transcription will still be activated by lactose.
D) All three genes will be expressed constitutively, regardless of the presence of lactose.
The Trp Operon
Bacteria use a five enzyme synthetic pathway to make the amino acid tryptophan from chorismic acid. In the presence of tryptophan, there is little point in making these enzymes, which are also co-localized in an operon.
The repressor protein is coded by the trpR gene (Figure 43). The repressor binds tryptophan when it is present, and the two together then bind the operator, to turn off transcription of the other five trp genes. In the absence of tryptophan, the bacterial cell must make its own. With no tryptophan present, the repressor protein cannot bind the operator. Without this block, RNA polymerase transcribes the five genes in the trp operon, and the five gene products allow the cell to make tryptophan. This is an example of anabolic repressible transcription.
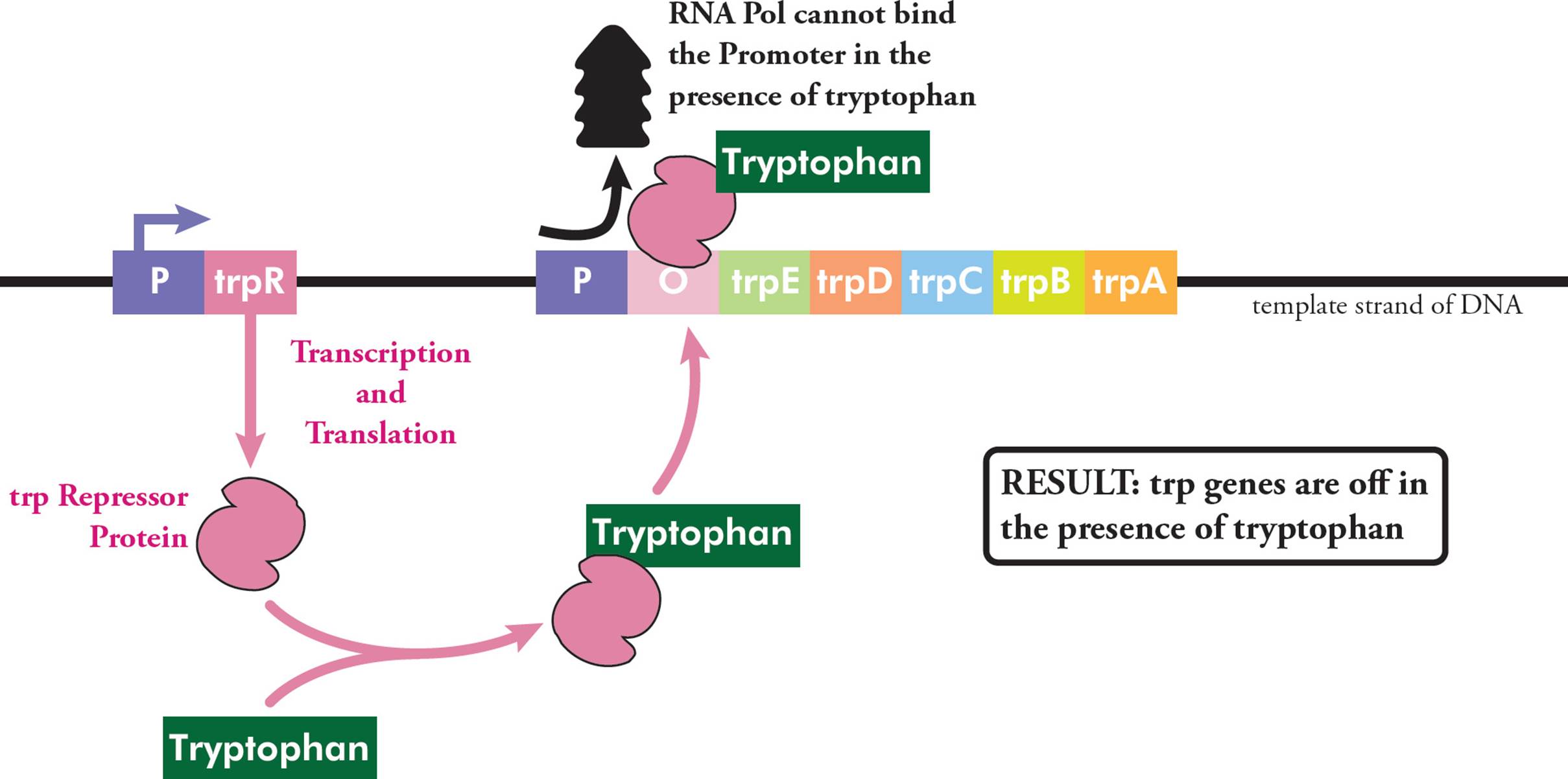
Figure 43 The Trp Operon in the Presence of Tryptophan
Regulation of Transcription: Eukaryotes
Given the complexity of eukaryotes compared to prokaryotes, it is not surprising that the regulation of eukaryotic transcription is also more complex. Most of this regulation happens at initiation.
For protein-coding genes, there are upstream control elements (UCEs), usually about 200 bases upstream of the initiation site, a core promoter containing binding sites for the basal transcription complex and RNA polymerase II (about 50 bases upstream of the transcription start site), and a TATA box at –25. The TATA box is a highly conserved DNA recognition sequence for the TATA box binding protein (TBP). Binding of TBP to the TATA box initiates transcription complex assembly at the promoter.
Enhancer sequences in DNA are bound by activator proteins, and this is another kind of transcriptional regulation. The enhancer may be located many thousands of base pairs away from a promoter (either upstream or downstream) and still regulate transcription. This is likely done by DNA looping so enhancers and their activator proteins can get close to transcriptional machinery.
Eukaryotes also have gene repressor proteins, which inhibit transcription; this can also be done by modifying chromatin structure. Transcription factors have DNA-binding domains and are crucial in transcription regulation. They can bind promoters or other regulatory sequences. In fact, in many cases, transcription levels in eukaryotes are controlled by huge committees of proteins. This produces a combinatorial effect, where each protein contributes to regulation, and can itself be regulated. These complex networks help link transcription to cell signaling and status. The binding of transcriptional machinery to DNA is often regulated by extracellular signals. For example, steroid hormones bind to receptors in the cell, and this sends the receptor to the nucleus. The complex binds DNA to regulate transcription. [If a mutation in a eukaryotic fat cell reduces the level of several proteins related to fat metabolism, does this mean the proteins are encoded by the same mRNA?75]
Beyond regulating the initiation of transcription, eukaryotes employ several other methods of transcriptional regulation, including:
• RNA Translocation: mRNA transcripts must be exported from the nucleus to the cytoplasm and can also be transported to different areas of the cell. They are translationally silent while this is happening. This system is especially important in cells that have a high level of polarity, where one area or end of the cell is distinctly different from the other. For example, neurons have polarity, and some transcripts are transported to the dendrites, while others stay in the soma. This is a way of controlling gene expression: mRNA transcripts aren’t translated into proteins until they are localized properly in the cell.
• mRNA Surveillance: Cells closely monitor mRNA molecules to ensure that only high quality mRNA transcripts are read by the ribosome. Defective transcripts (such as those with premature stop codons, or those without stop codons at all) and stalled transcripts (where the ribosome is stalled in translation) are degraded.
• RNA Interference: RNA interference (RNAi) is a way to silence gene expression after a transcript has been made. It is mediated by miRNA and siRNA (Section 5.7). Molecular biology labs often use the RNAi system experimentally, as a way to decrease protein expression (see Appendix I). Generally speaking, the siRNAs bind complementary sequences on mRNAs, and this ds-RNA is then degraded. The amount of transcript in the cell decreases, and gene expression is thus negatively regulated.
Translation Initiation
We’ve already discussed the complex process of assembling translational machinery. In both prokaryotes and eukaryotes, this is a highly regulated process, and links protein synthesis with upstream signaling pathways.
Post-Translational Modification
Newly synthesized proteins released from the ribosome are rarely able to function. They need to be correctly folded, modified or processed, and transported to where they function in the cell. These modifications are called post-translational events, since they occur after protein synthesis.
Protein Folding
First, the newly synthesized nascent protein is folded into its correct three-dimensional shape. This is accomplished by a family of proteins called chaperones. If folded correctly, the protein is said to be in its native conformation. If the protein is unfolded or misfolded, it’s said to be in its non-native state. Chaperone proteins are found across all types of organisms (from bacteria to plants to mammals), and also function in assembly or folding of other macromolecular structures. For example, chaperone proteins assist in nucleosome assembly from folded histones and DNA. In eukaryotic cells, chaperones are found in many subcellular compartments.
Covalent Modification
Many proteins are covalently modified. Some have hydrophobic groups added, to facilitate membrane localization. For example, the addition of a fatty acid can target a protein to a membrane (either the plasma membrane or an organelle membrane).
Smaller chemical groups can also be added. For example, proteins can be:
• Acetylated: addition of an acetyl group (–C(O)CH3), usually at the N-terminus of a protein, or at a lysine amino acid
• Formylated: addition of a formyl group (–C(O)H)
• Alkylated: addition of an alkyl group (such as methyl, ethyl, etc). Methylation is a common post-translational modification, and is usually done to lysine or arginine amino acids
• Glycosylated: addition of a glycosyl group to arginine, asparagine, cysteine, serine, threonine, tyrosine, or tryptophan amino acids. A glycosyl group is the substituent form of a cyclic mono- , di-, or oligosaccharide. This results in a glycoprotein.
• Phosphorylated: addition of a phosphate group (PO43–) to a serine, threonine, tyrosine, or histidine amino acid.
• Sulphated: addition of a sulphate group (SO42–) to a tyrosine amino acid.
Proteins can also be linked to other proteins. For example, in ubiquitination, proteins are covalently linked to ubiquitin.
There are many other examples of protein covalent modification. Overall, these modifications can have many effects on a protein and its function. They can change protein subcellular localization, target a protein for degradation, change interactions between proteins and other molecules, activate or inhibit enzyme activity, or change enzyme affinity for substrates. These modifications are typically studied in the lab using mass spectrometry (see MCAT Organic Chemistry Review, Chapter 5), western blotting, or eastern blotting (see Appendix I).
Processing
Many proteins require cleavage of some sort to become mature or functional. Cleavage can occur at either end of a peptide chain, or in the middle. Protein precursors are often used when the mature protein may be dangerous to the organism. Because the precursor is already made, it allows large quantity of mature protein to be available on short notice. Enzyme precursors are called zymogens or proenzymes.
A well-known example of post-translational processing is insulin. Insulin is made from a prohormone (Figure 44); preproinsulin is the primary translational product of the human INS gene. This peptide is 110 amino acids in length. To form proinsulin, an N-terminus signal peptide is removed and disulphide bonds form, in the endoplasmic reticulum. Three cleavage events are necessary to process proinsulin: the C peptide is removed by a family of enzymes called proprotein convertases, and a dipeptide fragment is removed from the C-terminus of the B chain peptide by a carboxypeptidase. These cleavage events occur in a secretory vesicle. The biological effects of insulin are well known, but it’s recently been shown that peptide C also has signaling properties.
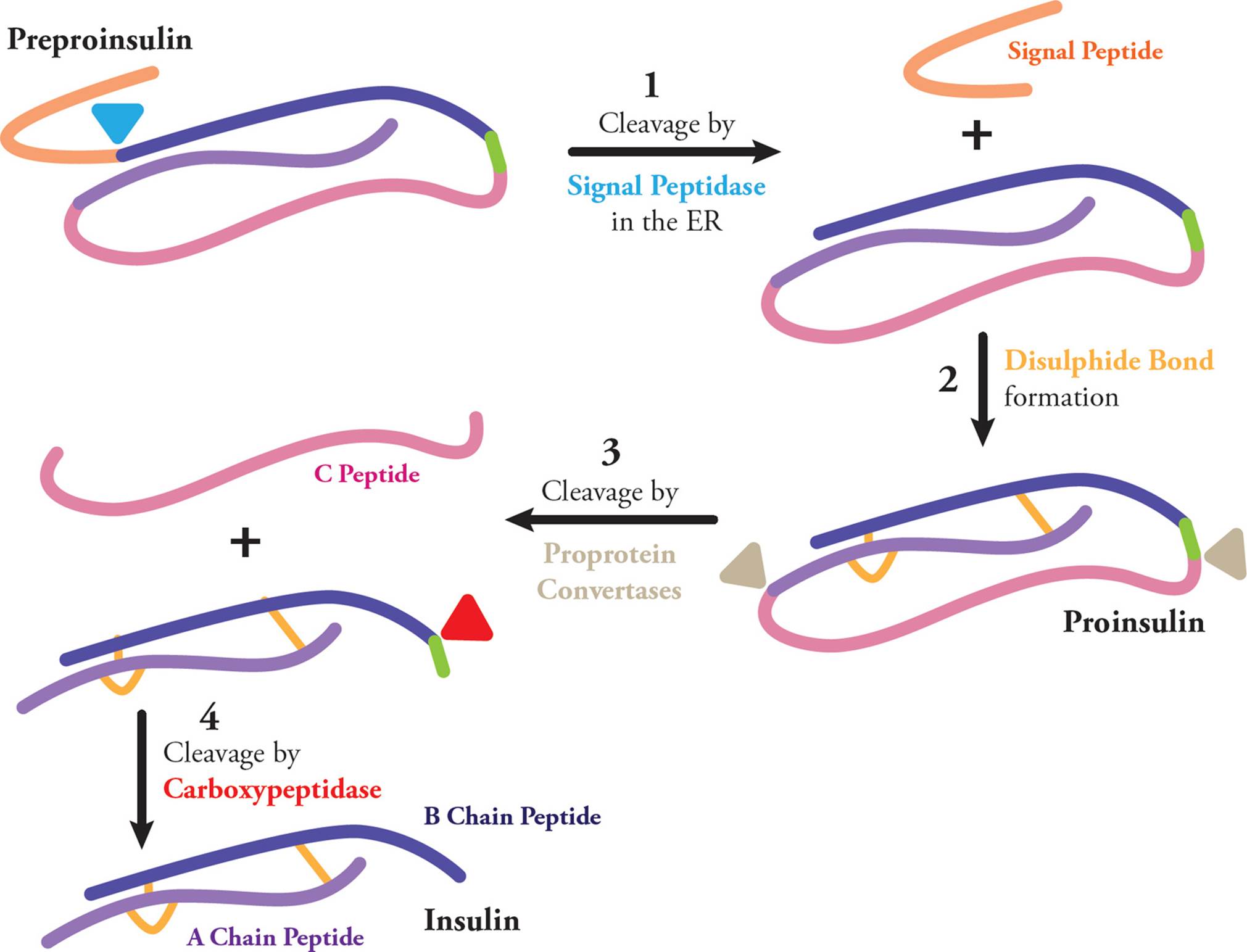
Figure 44 Insulin Processing: An Example of Post-Translational Modification
5.10 BEYOND NUCLEAR MOLECULAR BIOLOGY: ORGANELLE GENOMES
The endosymbiotic theory explains evolution of some eukaryotic organelles, such as mitochondria and chloroplasts. It suggests the origin of these organelles was as symbioses between separate single-celled organisms. The idea is that mitochondria and chloroplasts were each free-living bacteria that were individually taken inside a primitive eukaryotic cell as endosymbionts. The inner membrane of these organelles is derived from the former plasma membrane of these cells, and the outer membrane originated as the endocytosis vesicle when the bacteria were engulfed. This theory also explains why both these organelles contain additional genetic material; both the mitochondria and the chloroplast have their own genome, called mtDNA and ctDNA respectively. Both organelles are also capable of performing transcription and translation of this DNA, to make rRNAs, tRNAs, and proteins.
The human mitochondrial genome is small and only codes for 37 genes (Figure 45). Thirteen genes are protein coding, two code for rRNA, and the remaining 22 code for tRNAs. The rRNA and tRNA molecules are important because the mitochondrial genome uses a different genetic code (see Section 7.2) than the nuclear genome, and can perform its own transcription and translation. To give you a sense of scale here: mitochondria typically contain about 3000 different proteins, and only 13 are coded by mtDNA.
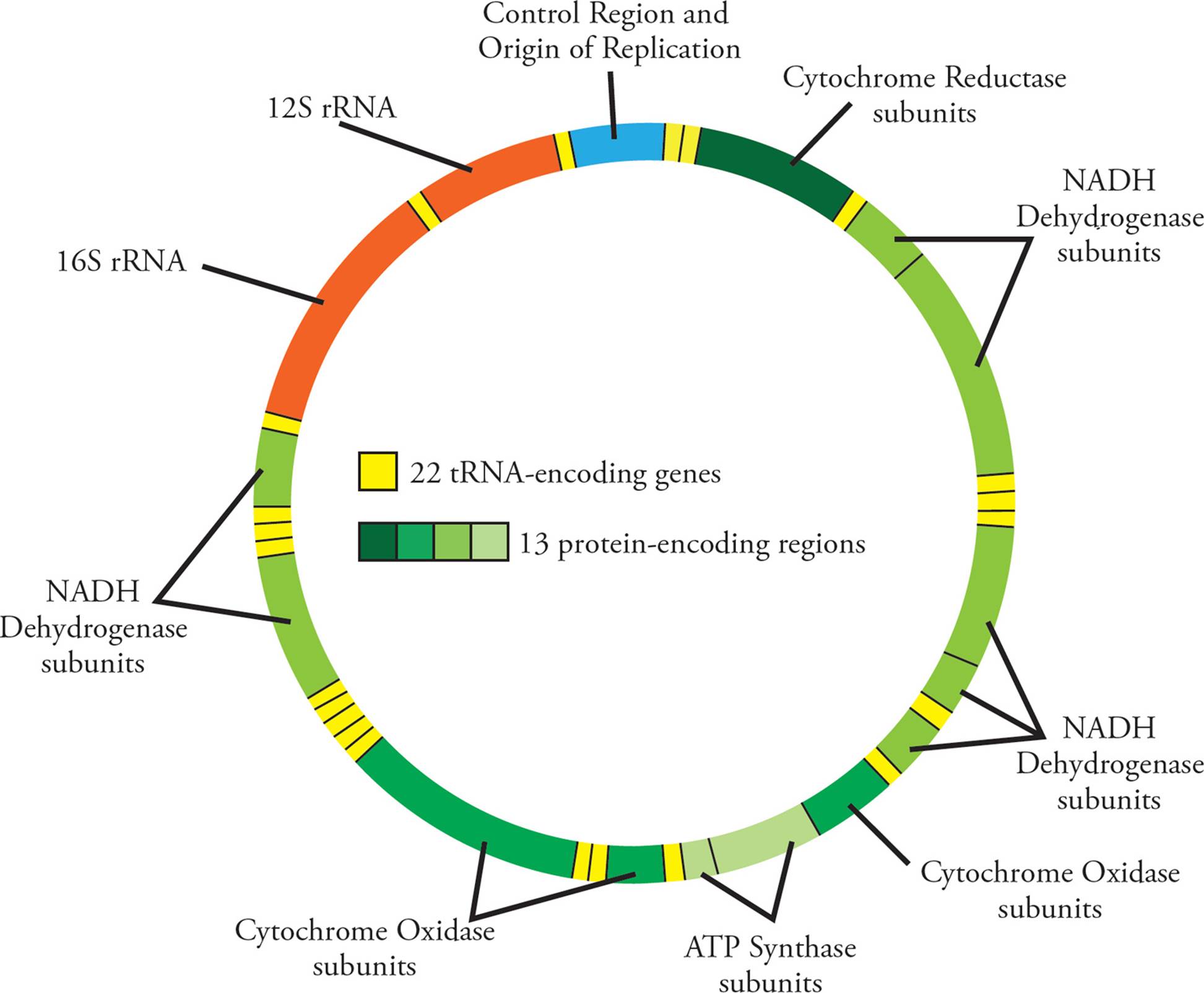
Figure 45 The Human Mitochondrial Genome
These 13 proteins are subunits of electron transport chain machinery, such as NADH dehydrogenase (complex 1), cytochrome c reductase (complex III), cytochrome c oxidase (complex IV), and ATP synthase. Each of these enzymes is quite complex with many subunits. For example, NADH dehydrogenase is made of 44 different peptide chains! mtDNA contributes to some of these subunits, but many also come from the nuclear genome.
One of the smallest prokaryotic genomes belongs to the bacteria Bartonella henselae, which makes about 1600 different proteins. If mitochondria evolved from prokaryotic organisms, why is the mitochondrial genome so small? There are two theories as to why this is the case. The first is that since the host cell already had a nuclear genome, there was little to no selective pressure for the mitochondrial genome to keep many protein-coding genes. Without any advantage to keeping these sequences, they were eventually lost. Second, many genes that were originally coded by mtDNA have since migrated to the nuclear genome. Old organelles are broken down by lysosomes, and this can liberate nucleic acid fragments, which then migrate to the nucleus.
Mutations in mtDNA can lead to phenotypes and traits, which often result in mitochondria-related diseases. Mitochondrial disorders are a heterogeneous group of diseases. They are often fatal at a young age, and are multi-systemic (meaning they affect many different systems of the body). Defects in oxidative phosphorylation are often the root cause of these diseases. About 15% of mitochondrial diseases are due to mutations in mtDNA. All humans receive mtDNA from their mother, as sperm contribute only nuclear DNA to a zygote. This means traits inherited via mtDNA display maternal inheritance, and this will be discussed more in Chapter 8.
mtDNA replication, transcription and translation are controlled by nuclear genes, even though the mitochondria codes for some of its own machinery. The mitochondrial genome has only three promoters and makes many polycistronic transcripts. Individual mitochondria can fuse and undergo exchange of genetic material, in a process that resembles conjugation. These characteristics display an evolutionary relationship to prokaryotic cells.
Each mitochondria contains an average of five copies of mtDNA, and there are about 100 mitochondria per cell. Even still, the amount of mtDNA in a eukaryotic cell is much less than the nuclear genome. mtDNA is commonly used in evolutionary biology and population history studies, because it has a high copy number and high mutation rate, doesn’t undergo recombination, and has maternal inheritance. The Y chromosome is also often used, for similar reasons (however, the Y chromosome is paternally inherited).
5.11 RETURN TO GENE STRUCTURE: A SUMMARY
Now that we have been through all the processes that a cell uses to turn a gene into a protein, and control this process, let’s review the components once again (Figure 46). Transcription begins at a start site, but needs a promoter upstream of this. It ends at a termination signal. The RNA transcript contains the open reading frame (which goes from start codon to stop codon), as well as both 5′ and 3′ regulatory regions.
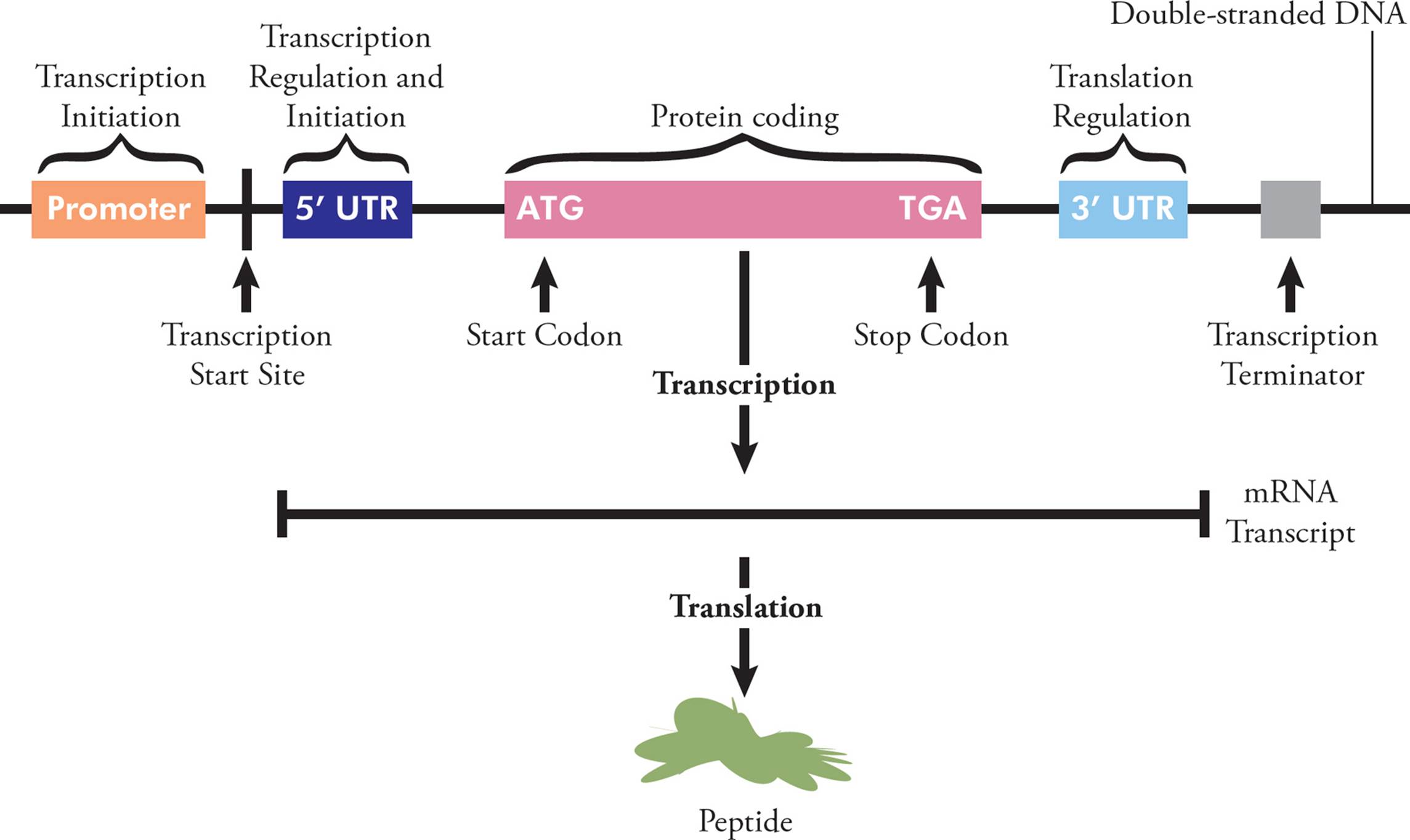
Figure 46 Gene Structure and Protein Expression
DNA replication, transcription, and translation have many similarities and some differences, and these are summarized in Table 1.

Table 1 A Review of Molecular Biology Processes
Chapter 5 Summary
• DNA is the fundamental unit of inheritance in cells.
• DNA and RNA are polymers, made of nucleotide monomers. A nucleotide contains phosphate group(s), a sugar (either deoxyribose for DNA or ribose for RNA), and a nitrogenous base, either a purine (adenine or guanine) or a pyrimidine (thymine, cytosine, or uracil).
• In DNA, adenine always pairs with thymine via two hydrogen bonds, and cytosine always pairs with guanine via three hydrogen bonds.
• Uracil replaces thymine in RNA, and the ribose in RNA has an OH group on carbon 2.
• DNA is supercoiled in prokaryotes and packaged around histone proteins in eukaryotes.
• Eukaryotic DNA is divided into several linear chromosomes which have unique structures including the long (q) and short (p) arms, centromere and telomeres on the ends.
• Genomes have extensive variation, including single nucleotide polymorphisms and copy number variation; transposons are mobile genetic elements which also contribute to genomic variation.
• Mutations can occur spontaneously, or can be caused by environmental factors (such as ionizing radiation), chemicals, or biological agents.
• Point mutations are classified based on their effect on the DNA (transition/transversion) or their effect on the amino acid sequence (missense, nonsense, or silent).
• Frameshift mutations are caused by insertions or deletions in the DNA base sequence that affect the reading frame of a gene. These are generally very serious mutations because they affect every amino acid codon from the point of the mutation on.
• Other types of mutations include inversions, translocations, and rearrangements.
• DNA replication occurs in the S-phase of the cell cycle and is semiconservative in nature.
• Several enzymes are involved in DNA replication. Helicases unwind the parental DNA at the origin of replication. Primases synthesize an RNA primer. DNA polymerase synthesizes new DNA, proofreads, and replaces the RNA primer. DNA ligase attaches the Okazaki fragments in the lagging strand.
• Cells have developed several ways to fix mutations, including: direct reversal, homology-dependent repair pathways (such as excision repair and post-replication repair), double-strand break repair (such as homologous recombination and nonhomologous end joining) and SOS repair.
• Transcription is the first part of protein synthesis; it is the creation of an RNA transcript by an RNA polymerase that reads the DNA template. Translation is the second part of protein synthesis; it is the creation of a polypeptide chain by ribosomes that read the mRNA transcript.
• There are several types of RNA that do not encode proteins. Some are directly involved in translation (rRNA and tRNA), while others play a role in gene expression (snRNA, miRNA, siRNA).
• Key info about Prokaryotes: theta replication, genome is a single circular piece of DNA, three different DNA polymerases, one RNA polymerase, no mRNA processing, polycistronic mRNA, simultaneous transcription/translation, smaller ribosomes.
• Key info about Eukaryotes: replication bubbles, genome is several linear pieces of DNA, one DNA polymerase, three RNA polymerases, capping, tailing, and splicing of mRNA prior to translation, monocistronic mRNA, transcription in nucleus, translation in cytosol, larger ribosomes.
CHAPTER 5 FREESTANDING PRACTICE QUESTIONS
1. A competitive inhibitor of eukaryotic RNA polymerase III would have the greatest effect on:
A) replication.
B) reverse transcription.
C) translation.
D) mutation.
2. In the lac operon, transcription is regulated by a repressor protein and only takes place in the presence of lactose. Which of the following statements is correct?
A) The repressor protein binds to the promoter site to inhibit transcription.
B) Lactose binds to the promoter site to initiate transcription.
C) Lactose binds to the repressor protein to inhibit transcription.
D) The repressor protein binds the operator site to inhibit transcription.
3. Which of the following could not be caused by a single point mutation in the DNA?
A) Ala-Gln-Cys-Asp-Leu → Ala-Gln
B) Ala-Gln-Cys-Asp-Leu → Ala-Gln-Cys-Asp-Leu
C) Ala-Gln-Cys-Asp-Leu → Ala-Gln-Cys-His-Lys
D) Ala-Gln-Cys-Asp-Leu → Ala-Gln-Cys-His-Leu
4. Which of the following is/are true with respect to eukaryotic mRNA?
I. Monocistronic
II. Transcription stops at the stop codon
III. Has the same sequence as the template DNA that it was transcribed from
A) I only
B) I and II
C) II and III
D) I, II and III
5. Which DNA base pair requires the most energy to break?
A) A-T
B) C-A
C) G-C
D) U-A
6. Which of the following is NOT a similarity between replication and transcription?
A) Both processes occur with the same fidelity.
B) Polymerization in both processes is based on reading a template.
C) A pyrophosphate is removed from every nucleotide as polymerization occurs.
D) Both processes occur in the 5 to 3 direction.
7. Telomeres are guanine-rich caps on the ends of each chromosome. Which of the following is the most likely function of a telomere?
A) High guanine content stabilizes parental strands to prevent excess tension during DNA unwinding.
B) Protect the ends of the chromosomes from damage due to incomplete replication
C) Provide a site for helicase attachment
D) Seal the gaps left by Okazaki fragments in the lagging strand
8. Which of the following functions is NOT typically attributed to small nuclear RNA (snRNA)?
A) Processing of pre-mRNA
B) Regulation of transcription factors
C) Coordinating amino acid addition in translation
D) Maintaining telomeres
9. Organisms with a higher degree of complexity do not necessarily have more diverse genomes than less complex organisms, in spite of the need for a greater diversity of proteins. Post-translational modification is one method used by more complex organisms to produce proteins that serve a wider variety of distinct functions. Which of the following explains this phenomenon?
A) The genome itself is manipulated by the agents responsible for post-translational modification in order to yield an increase in the number of transcriptional products.
B) Post-translational modification alters the structures and functions of proteins produced from a relatively smaller number of genes.
C) hnRNA is modified by the actions of post-transcriptional agents to provide increased variety in the mRNA used for translation.
D) Post-translational modifications enhance the ability of the ribosome to produce distinct protein products.
CHAPTER 5 PRACTICE PASSAGE
Protein synthesis involves a number of complex steps, from transcription of the gene through to translation and post-translational modification. After mRNA is transcribed in eukaryotes, it must be processed (capped, poly-A tailed, and spliced) before it can be translated. Prokaryotes do not need to process their mRNA.
Due to the exonuclease activity of DNA polymerase, DNA replication is generally a high-fidelity process. Random errors occasionally occur and these mutations are classified as frameshift mutations (insertions or deletions in the base sequence) or point mutations (a single base pair change). Any mutation is subject to natural selection, with advantageous mutations preserved and the most deleterious mutations eliminated quickly. Thus, areas of the genome that appear to evolve very slowly (i.e., have a slower rate of mutation than other areas) do not actually have a slower rate; rather, that area is highly critical to normal functioning of the organism involved.
Point mutations can be further classified by their final effect on the mature protein. Because of the redundancy of the genetic code, some mutations do not alter the final amino acid sequence of the protein and are referred to as silent mutations. However, it was discovered that all redundant codons are not equal; some are used preferentially to enhance the speed or accuracy of protein translation. tRNAs corresponding to redundant codons are not found equally in the cell; some tRNAs are more common than others. Silent mutations can cause phenotypic changes by altering mRNA stem-and-loop folding, half-life, and splicing sites. Thus, mutations formerly considered “silent” have now been implicated in a number of different disorders, such as Marfan syndrome, phenylketonuria, Seckel syndrome, and increased pain sensitivity.
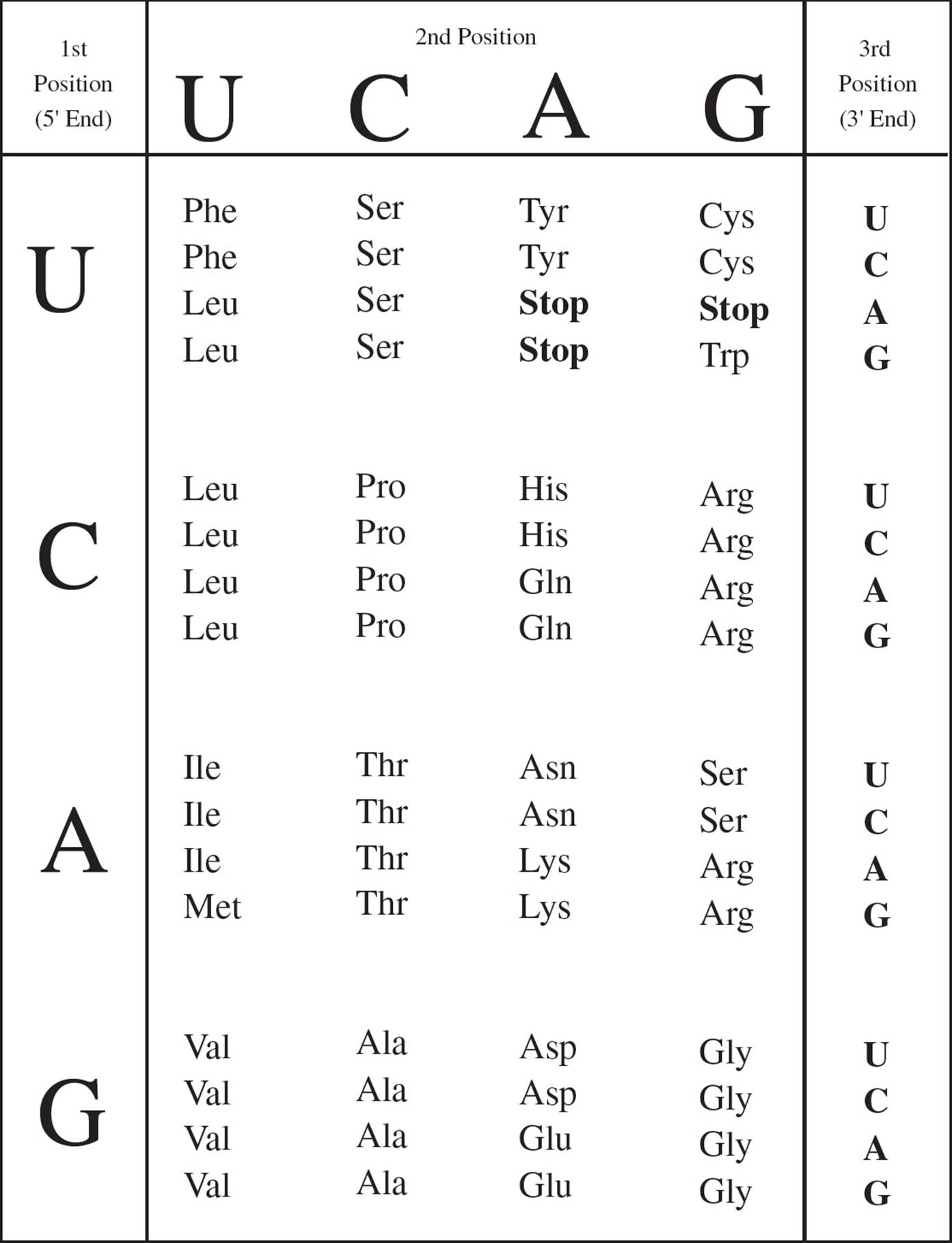
Figure 1 The Genetic Code
1. Based on information in the passage, genes coding for particularly abundant proteins in a cell would have all of the following EXCEPT:
A) codons corresponding to abundant tRNAs.
B) equal use of redundant codons.
C) greater use of preferential codons.
D) high-fidelity replication.
2. Which of following could account for the changes brought on by silent mutations in both eukaryotes and prokaryotes?
I. Decrease in mRNA half-life
II. Disruption of splicing sites
III. Changes in mRNA folding
A) I and II only
B) II and III only
C) I and III only
D) I, II, and III
3. Researchers studying a gene associated with breast cancer found that regions where silent mutations occur (“silent sites”) in this gene evolve very slowly compared to other regions within this gene. Comparisons were made between mice and humans. Which of the following is most likely true about this gene?
A) Mutations at other sites are more detrimental to the health of the organism than mutations at the silent sites.
B) Mutations at the silent sites increase the accuracy of mRNA splicing.
C) Mutations within the silent sites often lead to the death of the organism.
D) The silent sites are less critical to overall function than the other sites.
4. Which of the following has 3 to 5 exonuclease activity but NOT 5 to 3 exonuclease activity?
A) DNA pol III
B) DNA pol I
C) RNA pol II
D) RNA pol III
5. Point mutations are found in three subclasses: nonsense mutations, missense mutations, and silent mutations. Which of the following represents a silent mutation?
A) UGC to UGA
B) UUA to CUA
C) CAC to CAA
D) CAU to CUU
6. How could changing the half-life of an mRNA lead to phenotypic changes?
A) A shorter mRNA half-life would lead to a truncated protein.
B) A longer mRNA half-life would increase the amount of time the mRNA stays bound to the template strand of DNA, and reduce the amount of protein translated.
C) Differences in mRNA folding could alter the rates of translation.
D) More or less of the protein encoded by that mRNA would be translated.
SOLUTIONS TO CHAPTER 5 FREESTANDING PRACTICE QUESTIONS
1. C RNA polymerase III transcribes transfer RNA (tRNA), which then carries amino acids to ribosomes for use in translation. This polymerase plays no role in replication (choice A is wrong), and reverse transcription uses a DNA polymerase (in any case, it is not carried out by eukaryotes; choice B is wrong). Blocking the action of this enzyme would not alter the base sequence, so mutation would not be affected (choice D is wrong).
2. D The lac operon includes an operator site to which a repressor protein binds (choice A is wrong). The operator site is located between the promoter region and the start transcription site. When the repressor is bound, RNA polymerase (which binds to the promoter site; choice B is wrong) cannot move forward to the start site, thus transcription is inhibited (choice D is correct). Lactose binds to the repressor protein at an allosteric site, causing a conformational change so that the repressor protein can no longer bind to the operator. When this happens, RNA polymerase can move forward to the start site and transcription will occur (choice C is wrong).
3. C A point mutation is a single base pair substitution. There are few possibilities that can result if a single base is substituted. If the new codon is now a stop codon, then the polypeptide will be truncated (choice A could result from a point mutation and can be eliminated). If the new codon codes for the same amino acid as before the mutation, then a silent point mutation has occurred and no change will be seen in the amino acid sequence (choice B could result from a point mutation and can be eliminated). If the mutation leads to a single new amino acid, then a missense point mutation has occurred (choice D could result from a point mutation and can be eliminated). However, if more than one base was changed, or bases were added/deleted (a frameshift mutation), this would lead to multiple new amino acids (choice C could not result from a point mutation and is the correct answer choice).
4. A Item I is true: eukaryotic mRNA is monocistronic, meaning that only one protein is transcribed from each mRNA (choice C can be eliminated). Item II is false: Transcription does not stop at a stop codon; translation stops at a stop codon (choices B and D can be eliminated and choice A is the correct answer). Transcription stops when a termination signal is reached. Item III is also false: When mRNA is transcribed, it is complementary to the template strand, not identical to it.
5. C Guanine and cytosine base pairing involves three hydrogen bonds, whereas adenine and thymine only involves two. Therefore, G-C bonds would require more energy to break (choice A is wrong). Cytosine does not base pair with adenine (choice B is wrong), and uracil is an RNA base, not DNA base pairing (choice D is wrong).
6. A Fidelity refers to accuracy. Because RNA polymerases do not proofread, transcription is less accurate (i.e., a lower-fidelity process; choice A is not a similarity and is the correct answer choice). Both replication and transcription use DNA as a template (choice B is a similarity and can be eliminated). In both cases, the removal of pyrophosphate provides the energy for polymerization to occur (choice C is a similarity and can be eliminated). Lastly, although RNA polymerase (in transcription) and DNA polymerase (in replication) move along the parent chain in the 3′ → 5′ direction, the new chain is made in the 5′ → 3′ direction.
7. B Because DNA polymerase can only elongate DNA from a primer (i.e., a free 3′-OH group), the ends of the lagging strands do not get replicated. Even if an RNA primer bound to the very end of the chromosome, it would not be possible to replace the primer with DNA because there is no free 3′-OH group ahead of the primer to elongate. Thus, with each round of replication, the chromosomes get shorter. Since it is only a telomere at the end and not a critical gene, this can continue for several rounds of cell division, with the telomere getting shorter each time (choice B is correct). Ultimately, however, the telomere will get “used up,” and critical gene regions will begin to be shortened. At this point the cell enters senescence and is marked for destruction. Some cancer cells contain an enzyme (telomerase) that repairs the telomeres after replication, thus prolonging the cell’s life span. Topoisomerases help prevent excess tension, and in any case, high guanine content might make it more difficult for the parental strands to separate, leading to increased tension (choice A is wrong). Helicase binds at the origin of replication (choice C is wrong), and DNA ligase seals the gaps between Okazaki fragments (choice D is wrong).
8. C Transfer RNA (tRNA), not snRNA, is typically involved in the process of coordinating the amino acids that are added to a growing protein during translation (choice C is not a function of snRNA and is the correct answer choice). It should also be note that translation is NOT taking place in the nucleus, which is the location of snRNA. Processing of pre-mRNA, regulation of transcription factors, and maintenance of telomeres are all functions typically attributed to snRNA and take place in the nucleus (choices A, B, and D are all functions that include snRNAs and can be eliminated).
9. B Post-translational modification is, by definition, the manipulation of protein products after translation; the primary purpose of these modifications is to allow for an large increase in the number of possible protein products from a relatively small genome (choice B is correct). The genome, itself, is not changed during post-translational modification (choice A is wrong). hnRNA is processed in the nucleus after transcription to yield a variety of mRNA transcripts (and this is the other primary way that protein diversity can be achieved), but the question specifically asks about post-translational modification, not post transcriptional effects (choice C is wrong). The ribosome is essentially a factory that reads the code on an mRNA and links amino acids together, and that’s it. Ribosomes create the primary protein structure; this IS translation. Post-translational modification happens after this step (choice D is wrong).
SOLUTIONS TO CHAPTER 5 PRACTICE PASSAGE
1. B Proteins that are abundant require speed and accuracy during translation, and the passage states that this can be accomplished by using preferential codons (choice C is true and can be eliminated; choice B is false and the correct answer choice). Likewise, codons corresponding to abundant tRNAs would be used instead of those corresponding to the more rare tRNAs (choice A is true and can be eliminated). Choice D is true of all genes, abundant proteins or not (choice D can be eliminated).
2. C The passage states that silent mutations can lead to all three of the Roman numeral items listed; however, prokaryotes do not undergo mRNA splicing. Thus Item I is true for both eukaryotes and prokaryotes (choice B can be eliminated), Item II is only true for eukaryotes (choices A and D can be eliminated), and Item III is true for both.
3. C According to the passage, areas of the genome that appear to evolve very slowly are highly critical to normal functioning of the organism. Thus, mutations in these areas most likely disrupt function in a major way, leading to the death of the organism and thus the loss of the mutation (hence the reason it appears to evolve very slowly; choice C is correct and choice D is wrong). If other sites appear to evolve more quickly, mutations at those sites must be less detrimental (choice A is wrong). If mutations at the silent sites increase the accuracy of mRNA splicing, this would be beneficial, and thus preserved (choice B is wrong). Note that the information on breast cancer and humans vs. mice is not necessary to answer the question and is there solely to distract you. Focus on what the question is asking you.
4. A Enzymes with exonuclease activity can remove base pairs from the ends of nucleic acid strands. This can happen in either the 3 to 5 direction, or the 5 to 3 direction. This removal of nucleotides is necessary, for example, to fix polymerization mistakes (3 to 5), and also to remove the RNA primer during DNA replication (5 to 3). RNA polymerases do not correct their errors, so they have no exonuclease activity in either direction (this is a 50/50 question and now choices C and D can be eliminated). DNA pol III has only 3 to 5 error correction; as it is replicating DNA in the 5 to 3 direction, if it makes a mistake it can back up—3 to 5′—and correct its mistake (choice A is correct). DNA pol I has both exonuclease activities. If while synthesizing DNA it makes a mistake, it can back up and correct it (3 to 5 exonuclease activity), and it also has 5 to 3 exonuclease activity so that it can move in the 5 to 3 direction, remove the RNA primer, and replace it with DNA (choice B is wrong).
5. B Nonsense mutations convert a codon for an amino acid into a stop codon, missense mutations lead to amino acid substitutions, and silent mutations do not affect the amino acid sequence of a protein. To answer this question, you must use the genetic code in Figure 1. UGC codes for cysteine and UGA is a STOP codon, making this a nonsense mutation (choice A is wrong). The codons UUA and CUA both code for leucine, making this a silent mutation (choice B is correct). CAC codes for histidine and CAA codes for glutamine; this is a missense mutation (choice C is wrong). CAU codes for histidine and CUU codes for leucine; this is also a missense mutation (choice D is wrong).
6. D If the half-life of an mRNA is increased, it will stay in the cell longer and more of the protein would be translated. Likewise, if the mRNA’s half-life is decreased, it will be eliminated from the cell more quickly and less of the protein would be translated. The mRNA half-life has nothing to do with the length of the protein; protein size is dictated by the length of the open reading frame on the mRNA molecule and the number of codons in the translated region (choice A is wrong). mRNA does not stay bound to the DNA template strand for any length of time, regardless of half-life. As mRNA is transcribed, the DNA helix reforms immediately behind it, releasing the mRNA from the transcription bubble as it is synthesized (choice B is wrong). Choice C is a true statement but does not address the question of half-life (choice C is wrong).
1 The 2 OH is missing, so it is deoxyribose.
2 A mnemonic for this is: Pyramids (pyrimidines) have sharp edges, so they CUT. The U stands for uracil, which is a pyrimidine found in RNA instead of T. Another mnemonic is CUT the Py.
3 A beta linkage indicates that the anomeric carbon has a configuration with the attached group (a nitrogen of the aromatic ring of a purine or pyrimidine base) drawn above the plane of the ribose ring. Remember, it’s better to β up!
4 No. In dilute solution they will be H-bonded to water. However, H-bonds are the key determinant of the double-stranded structure of DNA; in DNA the bases do not interact with water because DNA coiling places them inside the tube-like structure of the double helix, where they interact with each other.
5 Peptide bonds with a carbon between them are the backbone, and the R group attached to the α carbon is the variable portion.
6 Since the backbone is the same regardless of the nucleotide sequence, the specificity in binding must be derived from interactions with bases.
7 During polymerization of nucleoside triphosphates, pyrophosphate is released and hydrolyzed, driving the polymerization reaction forward. Hydrolysis of the high energy pyrophosphate molecule makes the polymerization of nucleoside triphosphates more energetically favorable.
8 The T is written last and is therefore the 3 nucleotide, or the nucleotide with the free 3 hydroxy group.
9 Antiparallel H-bonding is reminiscent of the β-pleated sheet, which is a common secondary structure (it can be quaternary, when two separate chains come together to form a sheet).
10 This fact has a fringe benefit: We can calculate the number of purines if we know the number of pyrimidines. We can actually calculate several variables. Chargoff’s rule states that [A] = [T] and [G] = [C]; and [A] + [G] = [T] + [C].
11 The Tm of the first oligonucleotide pair would be lower because it contains more AT pairs. A and T only form two hydrogen bonds while G and C form three. Thus, it takes less kinetic energy to disrupt A-T rich ds-DNA than G-C rich ds-DNA.
12 The charged phosphates electrostatically repel each other in normal DNA. Methyl esters will not be charged. The lack of electrostatic repulsion between the methyl ester backbones will increase the Tm, meaning that more kinetic energy will be required to melt the oligonucleotides.
13 Item I: True. For every G, there is a C; and for every A there is a T. Item II: False. The ratio of purines to pyrimidines is always the same (50:50) since each purine is paired with a pyrimidine. In order to calculate the amount of any one base, you have to know the ratio of AT to GC pairs. Item III: False. Again, the ratio of purines to pyrimidines is always the same; 50:50. However, two chains containing mostly GC pairs will bond more tightly than two chains containing mostly AT pairs, since GC pairs are held together by 3 H-bonds while AT pairs have only 2. Item IV: True.Remember: the strands are antiparallel, A and T pair, G and C pair, and the 5 end is always written first.
14 Once a purine is H-bonded to a pyrimidine, most of the polar nature of the individual bases disappears because the charge dipoles are occupied in H-bonds.
15 Since one angstrom is 10−10 meter, the length is (3.4 × 10−10 meters/base pair)(9 × 107 base pairs) = 30 × 10−3 meters = 30 millimeters.
16 They’re mostly basic, since they must be attracted to the acidic exterior of the DNA double helix. This basicity is supplied by the amino acids arginine and lysine, which are unusually abundant in histones.
17 3 × 109 base pairs × 1 SNP/1000 base pairs = 3 × 106 SNPs, or approximately 3 million human SNPs.
18 Transformation and bacteriophage will be discussed in Chapter 6.
19 To transcribe a letter is to listen to spoken words and write them down as printed text. The message doesn’t change, and the language, English, doesn’t change. To translate a letter is to change it from one language to another. Cellular transcription is the process whereby a code is read from a nucleic acid (DNA) and written in the language of another nucleic acid (RNA), so the language is the same. In cellular translation, nucleic acids are read and polypeptides are written, so here the language does change.
20 With four nucleotides, if a “word” (codon) is two nucleotides long, there are 42 = 16 possible codons; too few to specify 20 unique amino acids. However, there are 43 = 64 possible 3-letter “words,” and 64 is more than enough different codons to specify 20 unique amino acids. Thus, three nucleotides is the minimum codon size.
21 RNA is the nucleic acid that actually encodes protein during translation. RNA has U instead of T.
22 The RNA codon transcribed from the DNA will be CAC, coding for histidine.
23 The RNA would have to be CCACCACCACCACCACCACCAC.… This would yield polyproline if read as CCA, CCA, CCA. But if it were read as CAC, CAC, CAC, it would give rise to polyhistidine. If it were read ACC, ACC, ACC, it would encode polythreonine.
24 No, since CUN codes for leucine, regardless of what N is. Notice that switching the 3rd nucleotide in the majority of codons will have no effect.
25 A template is something that is copied. The metal plates used in printing presses are an example.
26 complementary
27 Yes. Separating the strands requires the breaking of many H-bonds.
28 Imagine two long ropes wound around each other. What happens if you pull them apart in the middle?
29 If the daughter is made 5′ to 3′, and the two strands have to end up antiparallel, the template must be read 3′ to 5′.
30 No. The DNA strands are antiparallel, meaning that the upper strand would have to be extended in a 3 to 5 direction, which is impossible. Note that the phrase “in the presence of excess nucleotides” is extraneous. It just means there are plenty of building blocks around. Typical MCAT smokescreen.
31 Yes.
32 Exonuclease means “cutting a nucleic acid chain at the end”. An endonuclease will cut a polynucleotide acid chain in the middle of the chain, usually at a particular sequence. Two important types of endonucleases are: repair enzymes that remove chemically damaged DNA from the chain, and restriction enzymes, which are endonucleases found in bacteria. Their role is to destroy the DNA of infecting viruses, thus restricting the host range of the virus.
33 5′ to 3′ polymerase; remember, all polymerization is 5′ to 3′.
34 Yes. The 3′ to 5′ exonuclease activity is the polymerase’s way of editing its work. Without this editing function, many more point mutations would occur due to the incorporation of wrong nucleotides. The normal polymerase is remarkably adept at sensing correct base pairing and removing bases that don’t belong.
35 For example, substituting a small hydrophobe such as valine for another small hydrophobe like leucine will probably cause little disruption of protein structure. Another way of defining conservative mutations is that they cause changes in primary structure but do not affect secondary, tertiary, or quaternary structure.
36 No. If you insert or delete one whole codon or several whole codons, you add or remove amino acids to the polypeptide without changing the reading frame.
37 The original RNA codes for Met-Lys-Gly-Pro-Phe-Lys. After the insertion, the oligonucleotide will code for Met-Lys-Gly-Pro-Phe-Glu- Met-Thr. Note that this contains different amino acids and it’s longer. The extra length is due to the fact that a stop codon, UGA, changed by the frameshift.
38 By definition, all mutations involve a change in the genotype. Most mutations also cause a change in the phenotype, but in the case of conservative mutations it is a very subtle change that would be hard to detect.
39 Because a cell’s DNA is necessary for the cell’s entire life. RNA is a transient molecule which is transcribed, translated, and destroyed. As a matter of fact, the reason RNA contains uracil also has to do with the reduced need for fidelity in transcription as compared to replication. Without getting into the details, thymine is easier for DNA repair systems to work with, while uracil is much less energy-costly to make. So RNA has uracil, DNA has thymine.
40 All cells require RNA production, even if they are not growing, in order to continually replenish degraded RNA. RNA contains the bases cytosine, guanine, uracil and adenine, but only DNA contains thymine. Thus, if thymine production is blocked, only DNA replication will be inhibited and only rapidly dividing cells such as cancer cells will be affected. Unfortunately, some normal cells in the body normally divide a lot (such as lining cells of the gut and hair follicles), explaining the side effects of chemotherapy.
41 For instance, if five enzymes are necessary for the synthesis of a particular molecule, then all five enzymes might be encoded on a single piece of mRNA.
42 Each tRNA must recognize a codon on mRNA and respond by delivering the appropriate amino acid to the ribosome. There are 20 different amino acids, so there at least 20 different tRNAs. However, there are 61 possible codons, so there could be as many as 61 different tRNAs. The actual number is between 20 and 61, because the third nucleotide of the codon is often not needed for specificity of the amino acid.
43 complementary
44 5 to 3
45 No, RNA pol does not require a primer. Remember, the primer in replication is a piece of RNA, made by an RNA polymerase.
46 The virus will have a very high rate of mutation. It is a general law that most mutations are harmful. Hence, individual viruses will be far less likely to survive than organisms with DNA genomes. However, the high mutation rate will allow the entire species of virus to evolve very rapidly, making it very successful as a parasite (since it will evade host defense systems).
47 the origin
48 The DNA strand must be complementary to the first strand we discussed. So the sequence must be TTTTTTTTT. Hence the transcript will have to be AAAAAAAAA. Because AAA codes for lysine, the oligopeptide would be Lys-Lys-Lys.
49 No, it is accurate for prokaryotes only. In eukaryotes, the RNA transcript must be processed (spliced) and transported out of the nucleus before it can be translated. We will discuss this in depth later in the chapter.
50 It is made first, since transcription proceeds from 5 to 3.
51 Two conceivable reasons: 1) mRNA has a very short lifespan; it is degraded rapidly, and more must be made if the protein is still needed. Note that this is consistent with the idea that regulation of gene expression occurs primarily at the transcriptional level since this is more efficient. 2) Viruses may inject RNA into the cell. If it does not have the correct cap and tail modifications, exonucleases will destroy it.
52 Here is an example of the use of splicing for the regulation of gene expression. The piece of RNA must have been hnRNA. In the cell-free system it underwent differential splicing to produce one of two different mRNA molecules. Apparently, Polypeptide 1 came from an mRNA which had more material spliced out than the mRNA coding for Polypeptide 2.
53 No. They must be available for base pairing with the codon.
54 The anticodon is different for each of the different tRNA molecules. Part of the rest of the molecule varies from one tRNA to the next, but about 60 percent is constant. The amino acid binding site is always the same: CCA (at the 3 end of the tRNA molecule).
55 The easiest way to calculate this is to Figure out the probability of getting all amino acids in the protein correct, in other words, we must use the non-error rate for our calculation, not the error rate. If the error rate is 1/1000, then the non-error rate is 999/1000. The probability of having no errors is .999n, where n = the number of amino acid residues. In other words, a single amino acid has .999 probability, or 99.9% probability of being correct. Two amino acids correct in a row have a .999 × .999 probability (.9992), or .998, or 99.8% probability of happening. Continuing in this manner, a 500-amino acid protein has a .999500 probability of being entirely correct, or .606, approximately a 60% probability. Longer proteins have a higher chance of containing errors.
56 A single base change in the anticodon of the tRNA for arginine could cause it to recognize the codon for serine. If that happened in the mutant bacteria, problems might ensue, but one good result would be that the correct amino acid would be incorporated at the mutated site in hexokinase (choice A is correct; note that point mutations in tRNA genes are actually a common means of suppression in bacteria). Increasing the rate at which RNA polymerase recognizes the promoter might increase the rate of transcription, but would not fix a mutant enzyme (choice B is wrong), and a base pair deletion in the hexokinase gene would cause a frameshift mutation and a serious significant change in protein structure and function (choice C is wrong). A point mutation during transcription of a tRNA molecule might have a temporary effect on a single bacterium, but would not be passed on to its progeny; remember than only DNA mutations have lasting effects and errors made during transcription are generally insignificant (choice D is wrong).
57 A site, since this is where the next amino acid to be added must bind.
58 5 first, since the mRNA is made 5 end first. Transcription and translation go in the same direction on mRNA.
59 It does not always occur at the very end. You can deduce this from the fact that mRNA is polycistronic. If there are more than one translation start site on the mRNA, they can’t all be at the 5 end.
60 For example, streptomycin and tetracycline bind to the 30S subunit of the prokaryotic ribosome. Chloramphenicol and erythromycin bind to the 50S subunit.
61 This may seem odd, as ATP is normally the energy molecule. But a high energy phosphate is a high energy phosphate. Another example is the GTP produced in the Krebs cycle.
62 In fact, cells of our immune system release cytotoxins when they sniff out fMet, because this chemical is a sure sign that bacteria are busily translating.
63 Refer to the genetic code table. The codon for methionine is AUG; that’s the start codon. It only initiates translation when it is preceded by a Shine-Dalgarno sequence (prokaryotes).
64 The direction of synthesis is N → C, since the N of amino acid #2 binds to the C of #1. As the polypeptide elongates, its N terminus will come snaking out of the ribosome.
65 It must, if the tRNA remains H-bonded to the mRNA while moving to another spot in the ribosome.
66 Because the bond between each amino acid and its tRNA is a high energy bond whose hydrolysis drives peptide bond formation. Remember that the aminoacyl-tRNA bond was formed using the energy of two phosphate bonds from ATP.
67 There are two phosphate bonds hydrolyzed per amino acid to make the aminoacyl-tRNAs, or 100 for the 50 amino acid polypeptide. Two phosphate bonds are required for each elongation step, one for the entrance of each new aminoacyl-tRNA into the ribosomal A site and the other for translocation. Since there are 49 elongation steps for a 50-amino acid protein, 98 high energy bonds are hydrolyzed during elongation. Finally, one GTP is hydrolyzed during initiation to position the first tRNA and mRNA on the ribosome. Thus, a total of 199 high-energy bonds are required for the translation of a 50-amino acid protein. In other words, it costs approximately 4n high-energy bonds to make a peptide chain, where n is the number of amino acids in the chain.
68 In eukaryotes, the answer is: yes, always, because eukaryotic mRNA is monocistronic. In prokaryotes, however, different polypeptides may be translated from a single piece of mRNA, since prokaryotic mRNA is polycistronic.
69 In order for processes in eukaryotes to occur simultaneously, they must occur in the same compartment. Transcription and splicing both occur in the nucleus and could therefore occur simultaneously (choice B is correct). Translation occurs in the cytoplasm while transcription and splicing occur in the nucleus, thus translation cannot occur at the same time as either of these processes (choices A and C are wrong). mRNA degradation and transcription cannot occur at the same time; if this were true no mRNA molecules would survive to be translated (choice D is wrong).
70 A quick glance at the data indicates that transcription is increased in the presence of the sequence specific factor (compare lines 1 and 2 with lines 3 and 4, choice B is wrong), and that histones decrease the rate of transcription (packaged DNA has a lower rate of transcription than unpackaged, choice C is wrong). Looking closer, it appears that the sequence specific factor causes an approximate 8-fold increase in the transcription rate of packaged DNA (compare lines 2 and 4), but doesn’t even double the rate of transcription of unpackaged DNA (compare lines 1 and 3). It might be that this occurs because the factor increases the rate of transition to an open complex, but there is no data to support this (choice A is a better answer than choice D). Don’t confuse “open complex” (which means separated DNA strands) with “unpackaged” (which means not wrapped around histones).
71 It takes a great deal of ATP to synthesize RNA and protein, so it’s more energy efficient to transcribe and translate only the proteins that are needed.
72 They are not synonymous. All polypeptides are gene products, but some gene products are not polypeptides and some polypeptides are not enzymes. Transfer RNA and rRNA are gene products, but not polypeptides. Microfilaments and other elements of the cytoskeleton, as well as collagen and many other polypeptides, are not enzymes.
73 So note: The default for repressible systems is “ON”; for inducible systems the default is “OFF.”
74 If the repressor cannot bind to the operator, nothing will prevent RNA polymerase from transcribing all the genes on the operon in an unregulated, constitutive (or continuous) fashion (choice D is true and choice B is false). All genes on the operon are expressed or repressed together (choice A is false), and lactose will no longer have any effect (the expression of the genes is unregulated, so choice C is false).
75 No, it does not. Eukaryotic mRNA is monocistronic. A more likely explanation is that a number of different genes located throughout the genome have related regulatory sequences that bind the same sequence-specific transcription factors. This is the means used by eukaryotes to achieve coordinated expression of genes. Related proteins are clumped together on the same piece of mRNA in prokaryotes only.