Biology Premium, 2024: 5 Practice Tests + Comprehensive Review + Online Practice - Wuerth M. 2023
UNIT 6 Gene Expression and Regulation
16 Transcription and Translation
Learning Objectives
In this chapter, you will learn:
➜Transcription in Prokaryotes vs. Transcription in Eukaryotes
➜Translation
➜Flow of Information from the Nucleus to the Cell Membrane
Overview
The term central dogma describes the typical flow of genetic information in a cell: DNA is transcribed into mRNA; mRNA is then translated into proteins by ribosomes. This chapter will review the processes of transcription and translation and explore how these processes are similar or different in prokaryotes and eukaryotes.
Transcription in Prokaryotes vs. Transcription in Eukaryotes
Transcription is the process in which the genetic information in a sequence of DNA nucleotides is copied into newly synthesized RNA molecules. RNA polymerase is the enzyme that transcribes a DNA sequence into RNA molecules. The function of an RNA molecule depends on its structure and sequence:
§ mRNA (messenger RNA) is single-stranded and carries information from DNA to the ribosome. mRNA contains three base pair sequences called codons, which are complementary to the DNA base pair sequence. These codons will specify specific amino acids during translation.
§ tRNA (transfer RNA) folds into a three-dimensional structure that acts as an adapter molecule during translation. One end of the tRNA will bind to a specific amino acid, while the other end of tRNA contains an anticodon, which will pair with the appropriate mRNA codon at the ribosome during translation.
§ rRNA (ribosomal RNA) also folds into a three-dimensional structure. rRNA and proteins form ribosomes that perform translation. The three-dimensional rRNA acts as a ribozyme, catalyzing the reactions needed in translation.
In music, transcription means the arrangement of a piece of music for different instruments. Think of transcription in biology as the arrangement of the “music” of genetic information in DNA for the different “instruments” of mRNA, tRNA, and rRNA.
To start transcription, the enzyme RNA polymerase must bind to a noncoding DNA sequence called a promoter. The promoter sequence does not code for any amino acids and instead serves as a binding site for RNA polymerase upstream from the start of the coding region of a gene. Promoter sequences are highly conserved in living organisms. Most eukaryotic promoters contain a region called a TATA box, so named because it is rich in thymine and adenine nucleotides. Proteins called transcription factors help RNA polymerase bind to the promoter sequence and begin transcription.
During transcription, RNA polymerase adds new RNA nucleotides in the 5′ to 3′ direction, similar to how DNA polymerase adds new DNA nucleotides in the 5′ to 3′ direction during DNA replication. The strand of DNA being transcribed by RNA polymerase is called the template strand. The newly synthesized RNA must be antiparallel to the template DNA strand, so RNA polymerase reads the template DNA strand in the 3′ to 5′ direction. The sequence of base pairs in the RNA strand that is transcribed is complementary to the template DNA sequence. For example, if the DNA sequence is 3′ — ACG TAC GTA CGT — 5′, the newly synthesized RNA sequence will be 5′ — UGC AUG CAU GCA — 3′. The template strand of DNA is also known as the “minus strand” or the “noncoding strand” or the “antisense strand.” This is because the sequence of the mRNA strand that will be read by the ribosome during translation is actually not identical to the sequence of the DNA strand from which it was transcribed but instead is the complement (or opposite) of the DNA strand from which it was transcribed. The strand of the double helix that functions as the template strand can vary depending on the gene being transcribed.
TIP
Remember that RNA polymerase is the enzyme responsible for transcription and forms RNA molecules while DNA polymerase is the enzyme responsible for DNA replication and forms DNA molecules. Be sure not to confuse these two enzymes on the AP exam.
Remember that one of the differences between prokaryotic and eukaryotic cells is that prokaryotic cells do not have a nucleus. Therefore, in most prokaryotic cells, the mRNA transcript formed in transcription is immediately accessible to ribosomes and can be translated without delay. In eukaryotic cells, the initial mRNA transcript, referred to as pre-mRNA, needs to be modified before it can leave the nucleus of the cell and travel to the ribosomes for translation.
In eukaryotic cells, three modifications must occur to the pre-mRNA before it can leave the nucleus: the removal of introns and the joining of exons, the addition of a guanosine triphosphate (GTP) cap to the 5′ end of the RNA, and the addition of a poly-adenine (poly-A) tail to the 3′ end of the RNA.
1.Eukaryotic pre-mRNAs contain noncoding RNA sequences called introns. These introns are interspersed between coding sequences of eukaryotic RNAs called exons. Structures called spliceosomes, which are made of small nuclear RNAs (snRNAs) and small nuclear ribonucleoproteins (snRNPs), remove the introns from the pre-mRNA and then splice together the exons. These exons can be joined in different combinations to generate multiple RNA transcripts from the same gene, as shown in Figure 16.1. This alternative splicing gives eukaryotes the ability to generate a greater variety of RNA transcripts from just one gene than what can be generated from one gene in prokaryotes.
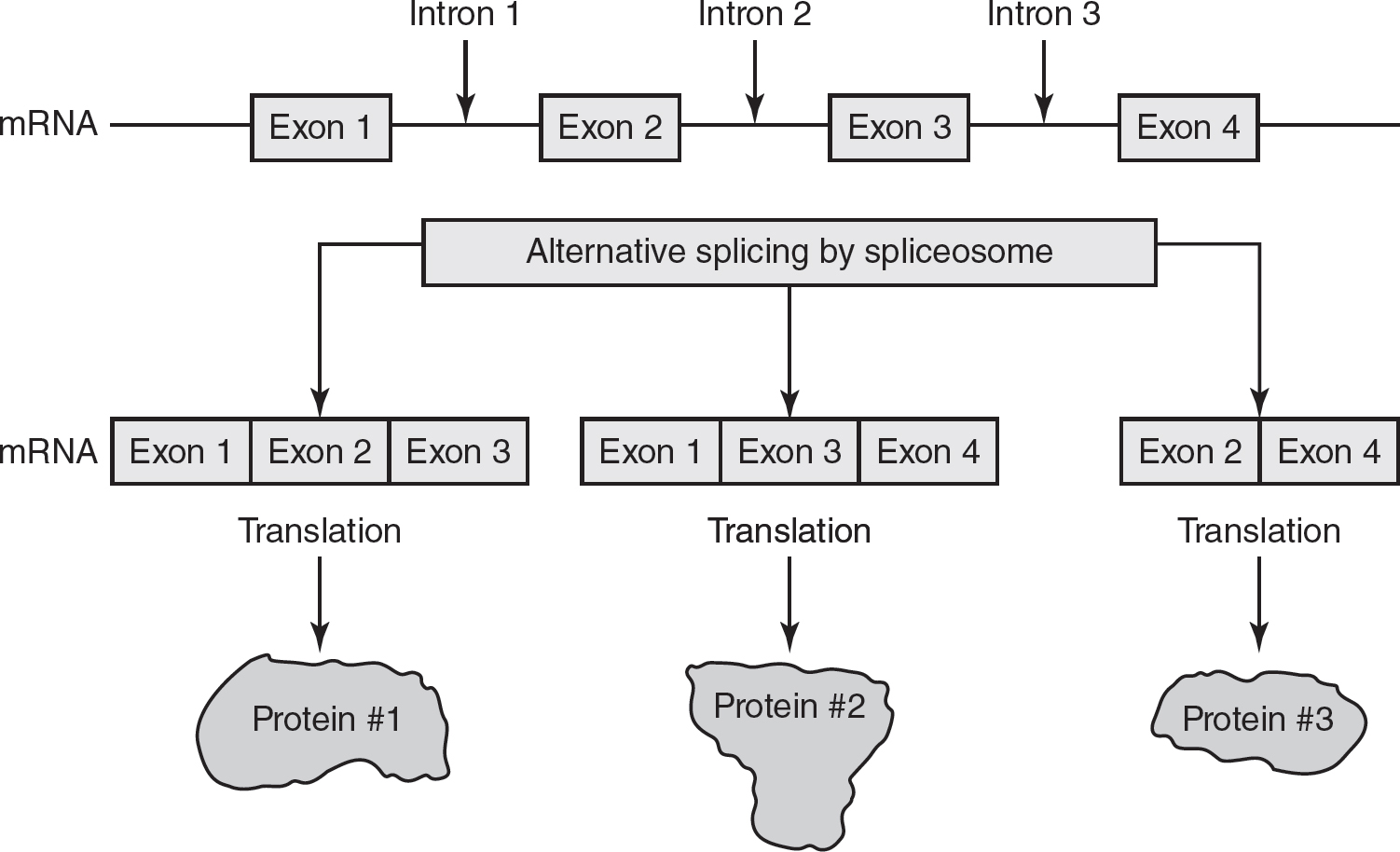
Figure 16.1 Alternative Splicing
While introns do not code for amino acids, some introns may function in the regulation of gene expression.
2.To protect the 5′ end of the pre-mRNA transcript from degradation before it can be translated, a 5′ GTP cap is added. Nuclear pores recognize this 5′ GTP cap and allow mRNAs with the cap to exit the nucleus. This GTP cap also helps in the initiation of translation when the RNA reaches the ribosome.
3.The enzyme poly-A polymerase adds a string of adenine nucleotides to the 3′ end of the pre-mRNA transcript. This 3′ poly-A tail helps prevent degradation of the transcript. mRNAs with longer 3′ poly-A tails tend to have longer durations in the cytosol, which allows more copies of the protein (that the mRNA codes for) to be generated.
After the excision of the introns and splicing of the exons, as well as the additions of the 5′ GTP cap and the 3′ poly-A tail, the transcript is now referred to as a mature mRNA and is ready to be translated by the ribosome.
Translation
Translation of mRNA molecules occurs at the ribosomes in both prokaryotes and eukaryotes. Ribosomes are found in the cytoplasm of both prokaryotes and eukaryotes. Ribosomes are also found on the rough endoplasmic reticulum of eukaryotes. In eukaryotes, cytoplasmic ribosomes usually translate proteins that will stay inside the cell, while ribosomes on the rough endoplasmic reticulum usually translate proteins that will be exported from the cell.
In the human immune system, alternative splicing of exons in the antibody-producing B cells allows for the production of a wide variety of antibody molecules in response to evolving pathogens.
Because prokaryotes do not have a nucleus, translation of the mRNA can occur as the mRNA is being transcribed. Multiple ribosomes can be simultaneously translating a prokaryotic RNA, forming polyribosomes (also known as polysomes), as shown in Figure 16.2.
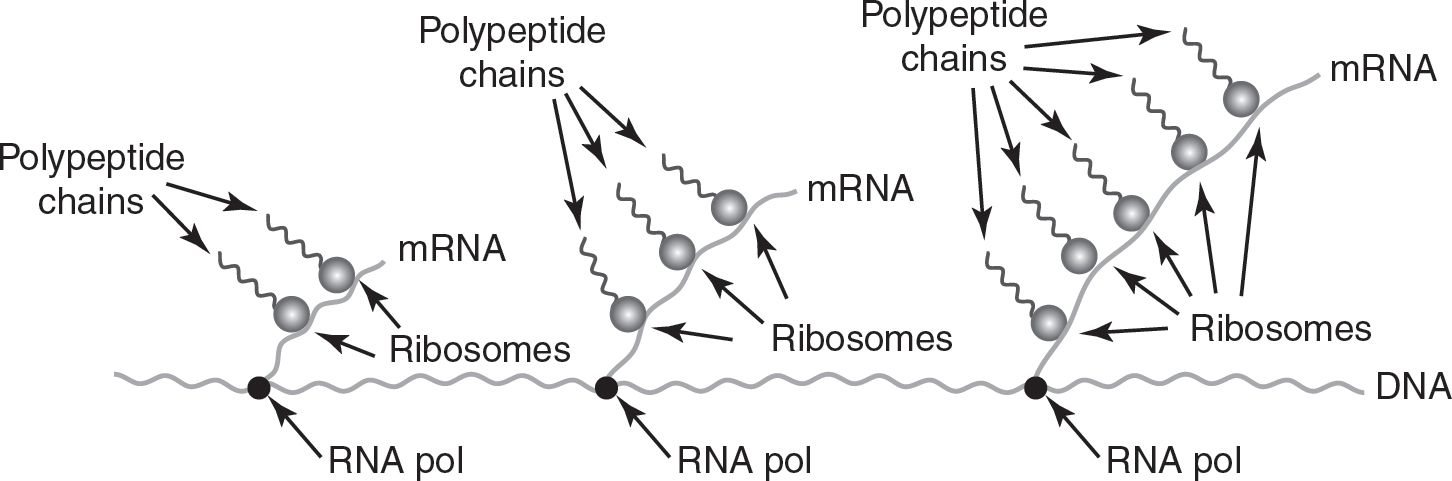
Figure 16.2 Polyribosomes
The process of translation requires energy and involves three main steps in both prokaryotes and eukaryotes: initiation of translation, elongation of the polypeptide chain, and termination of translation.
1.Initiation: The genetic code in mRNA is read in three base pair units called codons. Translation is initiated when the rRNA in the ribosome pairs with the start codon (AUG). A tRNA with the complementary anticodon (in this example, UAC) brings the appropriate amino acid to the ribosome, and the anticodon on tRNA pairs with the codon on the mRNA. In this way, tRNA functions as an “adapter” molecule, linking the correct amino acid with the correct codon on the mRNA. This process is called initiation and is shown in Figure 16.3.
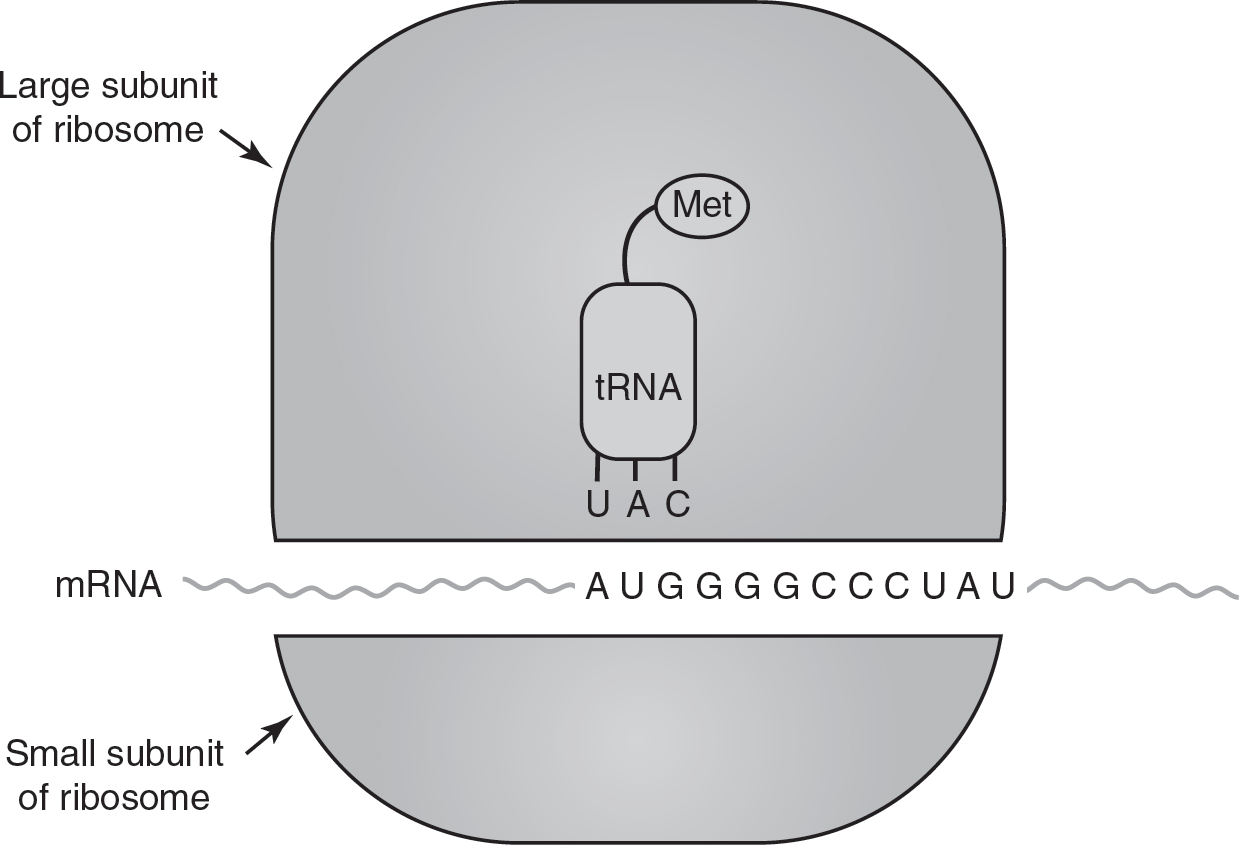
Figure 16.3 Initiation of Translation
Each codon on mRNA codes for one amino acid, but multiple codons can code for the same amino acid. For example, the codons UCA, UCC, UCG, and UCU all code for the amino acid serine. This gives redundancy to the genetic code. This redundancy in the genetic code may result in “silent” mutations that do not affect the amino acid sequence of the polypeptide chain. Almost all organisms on Earth use the same genetic code, which is evidence for the common ancestry of all organisms.
As Figure 16.4 shows, 61 of the 64 possible codons code for amino acids. Three of the 64 codons do not code for amino acids and are stop codons.
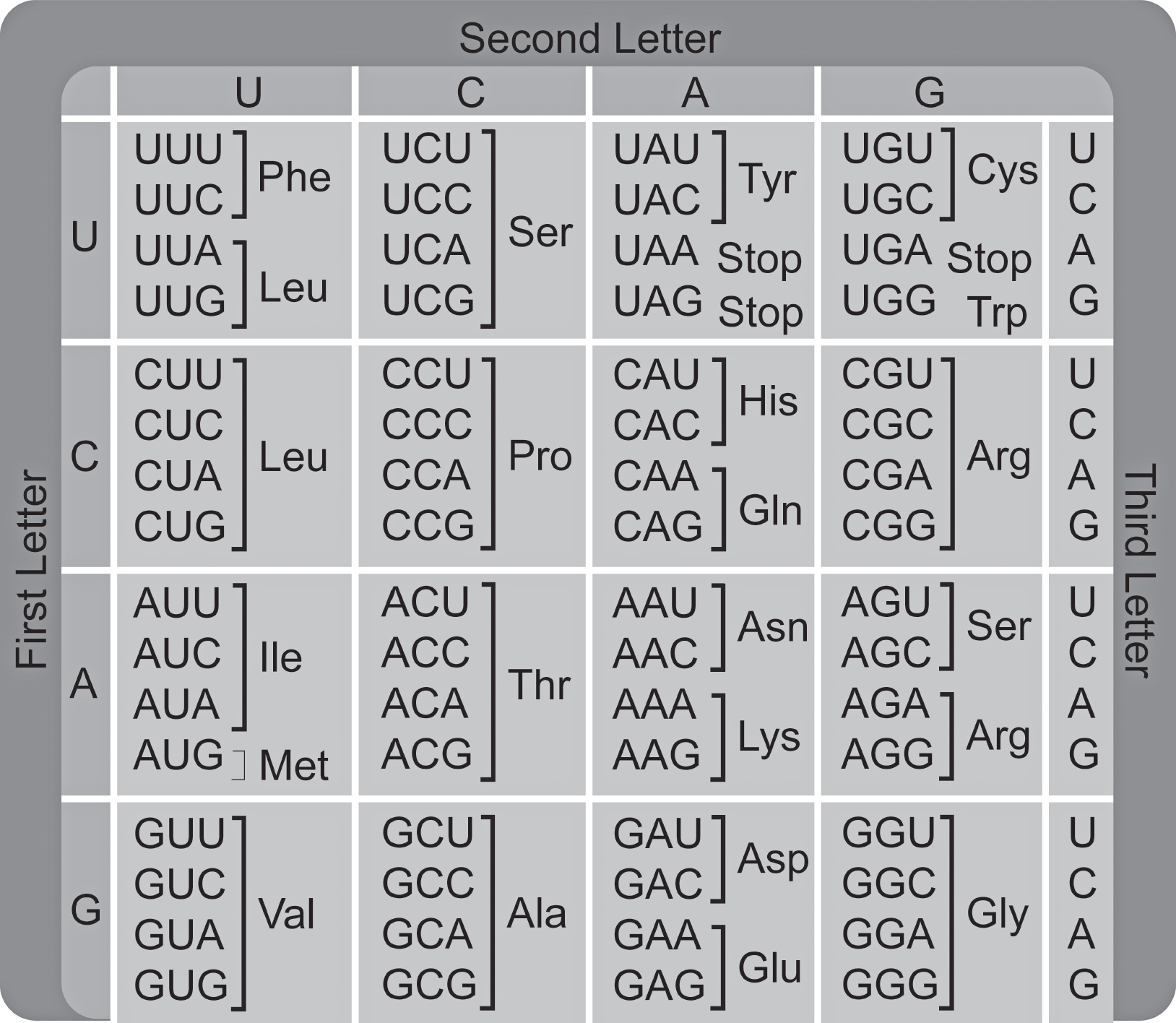
Figure 16.4 The Genetic Code
TIP
You do not need to memorize any codons of the genetic code. If needed, a codon chart will be provided on the AP Biology exam. Codons are specified here for illustrative purposes and clarity.
2.Elongation: After the first amino acid is placed in the ribosome by tRNA, the ribosome translocates (or moves) to the next codon. A new tRNA with the appropriate anticodon and amino acid then pairs with this codon. The ribosome then catalyzes the formation of a peptide bond between the amino acids brought to the ribosome by the first two tRNAs, forming the beginning of the polypeptide chain. Once the peptide bond is formed between the amino acids, the first tRNA releases its amino acid (which is now linked to the second amino acid), and the first tRNA is released from the ribosome. (See Figure 16.5.) This translocation of the ribosome and the addition of new amino acids to the polypeptide chain is called elongation and is repeated one codon at a time until a stop codon is reached.
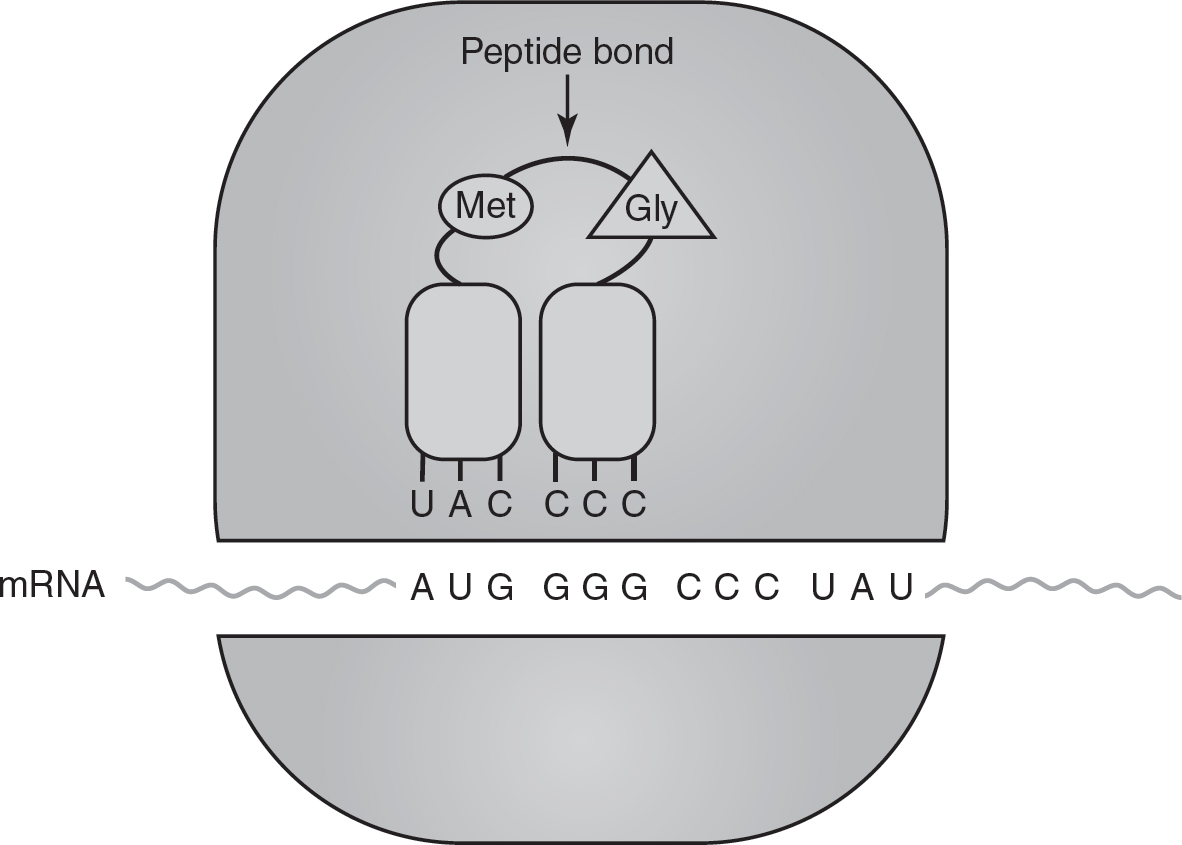
Figure 16.5 Elongation of Translation
3.Termination: Stop codons (also known as “nonsense” codons) do not code for any amino acid. As shown in Figure 16.6, when the ribosome reaches a stop codon, proteins called release factors bind to the ribosome. These release factors cause the ribosome to disassemble and release the polypeptide chain. This ends translation and is called termination.
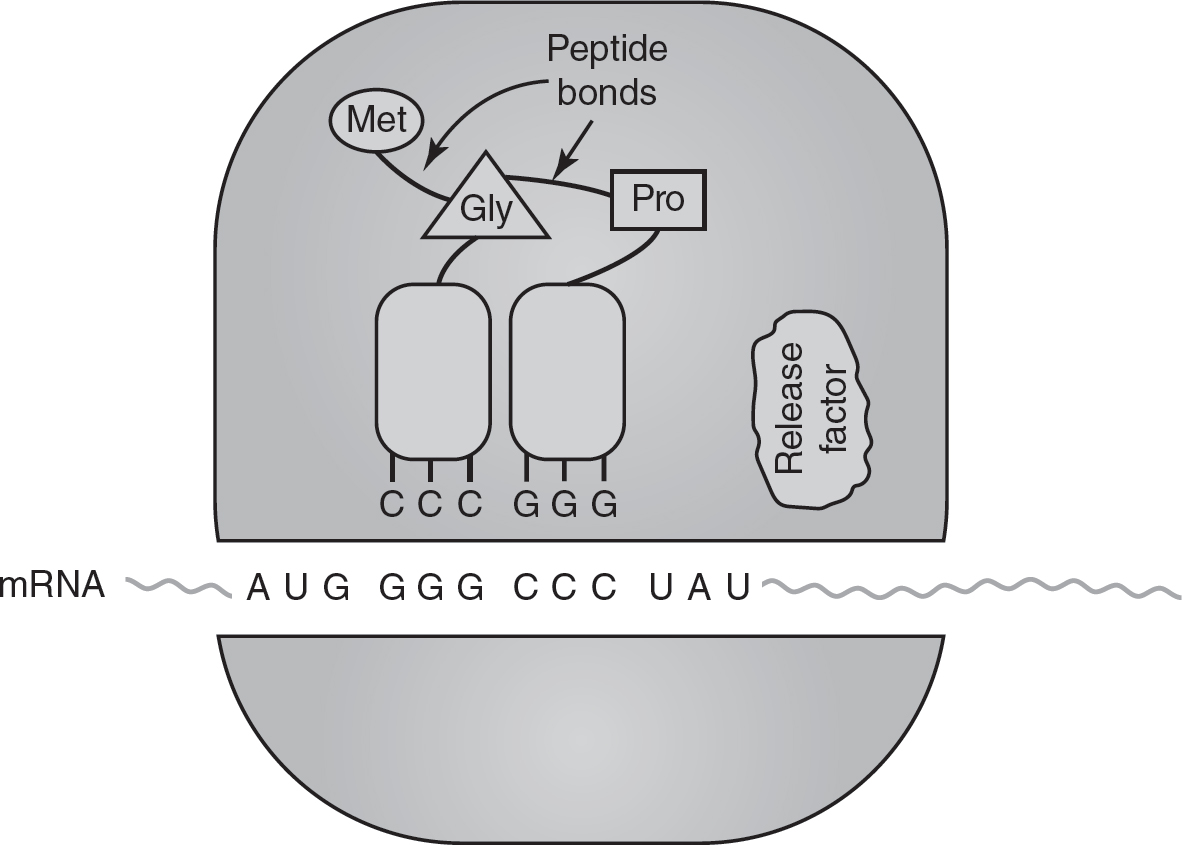
Figure 16.6 Termination of Translation
Flow of Information from the Nucleus to the Cell Membrane
DNA and RNA are the carriers of genetic information in organisms. In most organisms, genetic information starts with DNA, which provides the information for the transcription of mRNA. mRNA then provides the information for the sequence of amino acids in a protein.
In eukaryotes, genetic information in DNA is transcribed into mRNA in the nucleus. Ribosomes on the rough endoplasmic reticulum use the information in mRNA to translate proteins. After the protein is translated, a vesicle containing the protein will bud off from the rough endoplasmic reticulum and travel to the Golgi. At the Golgi, the proteins will be modified and packaged into vesicles for export from the cell. These vesicles will bud off from the Golgi and travel to the cell membrane. The vesicles then fuse with the cell membrane and release their protein contents from the cell. So the flow of genetic information in a eukaryotic cell is as follows:
DNA → mRNA → protein at ribosomes on the rough endoplasmic reticulum → protein at the Golgi → protein in vesicle → protein outside of cell membrane
This flow of genetic information is represented in Figure 16.7.
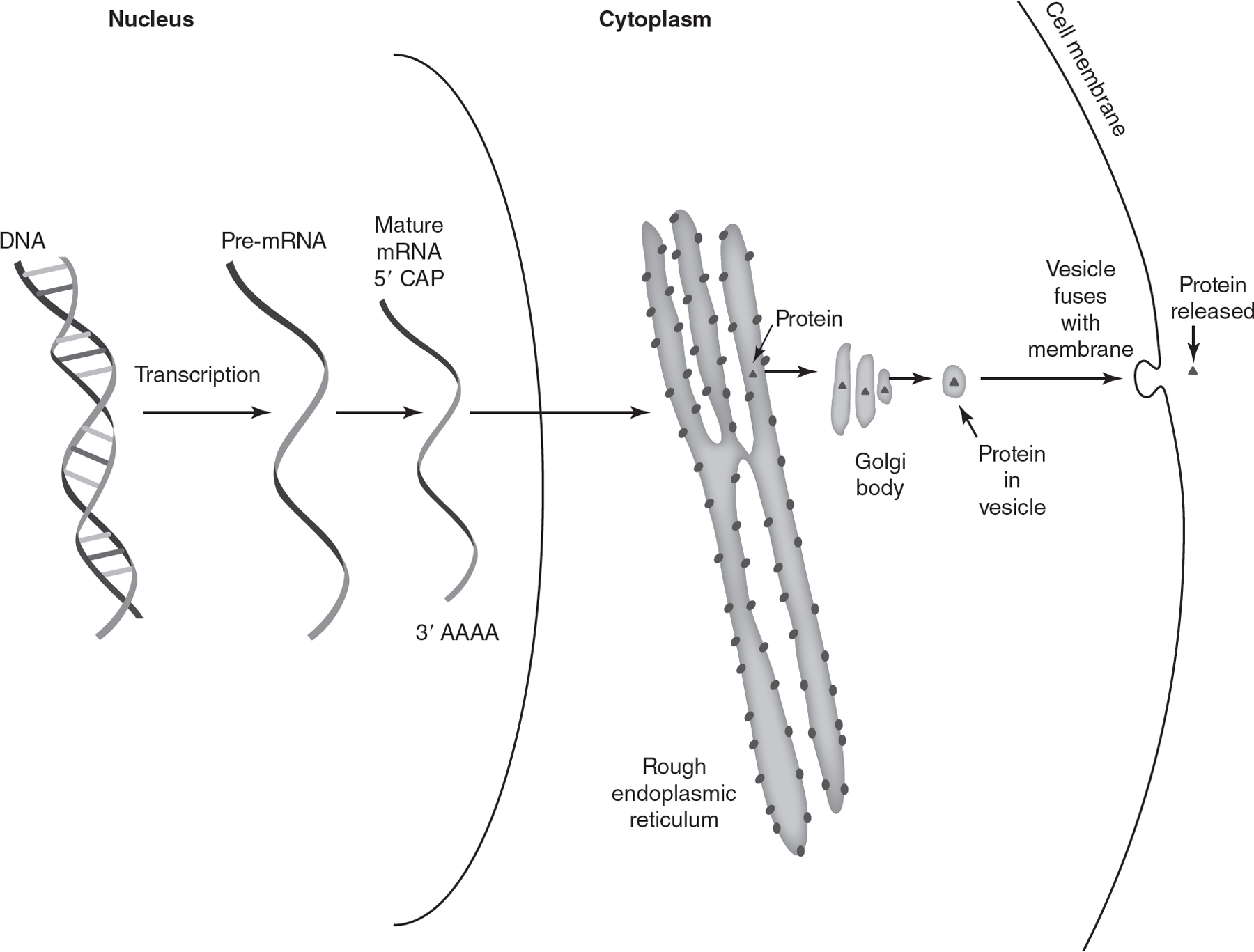
Figure 16.7 Flow of Genetic Information in a Eukaryotic Cell
Some viruses contain RNA as their primary carrier of genetic information. These retroviruses contain the enzyme reverse transcriptase. Reverse transcriptase makes a DNA copy of the RNA genome of the virus. This DNA copy is then inserted into the genome of the host cell that is infected by the virus. The host cell will then transcribe and translate the information in the viral DNA inserted into the host cell’s genome. Reverse transcriptase is less accurate than RNA polymerase, so retroviruses have a relatively high mutation rate.
Practice Questions
Multiple-Choice
1.What would be the minimum number of nucleotides required to code for a protein made of 12 amino acids?
(A)6
(B)12
(C)36
(D)48
2.A scientist inserts a eukaryotic gene directly into a bacteria’s genome. However, the protein produced by the bacteria from the eukaryotic gene does not have the same amino acid sequence as the protein produced from the gene in eukaryotic cells. Which of the following best explains this?
(A)Prokaryotes and eukaryotes do not use the same genetic code, so the bacteria cannot decode the eukaryotic gene.
(B)Eukaryotic genes contain introns, which must be removed before translation; prokaryotes cannot remove introns.
(C)Prokaryotes do not have a nucleus and cannot perform transcription.
(D)Prokaryotes do not have rough endoplasmic reticulum, so they cannot translate eukaryotic genes.
3.Which of the following correctly represents the mRNA sequence that would be transcribed from the DNA sequence 3′ ACC GGT AAG TTC 5′?
(A)3′ TGG CCA TTC AAG 5′
(B)3′ UGG CCA UUC AAG 5′
(C)5′ TGG CCA TTC AAG 3′
(D)5′ UGG CCA UUC AAG 3′
4.What is the amino acid sequence that would be translated from the following gene? (Use Figure 16.4 to answer this question.)
3′ AAT CGT TTC AAT CAA 5′
(A)Asn-Arg-Phe-Asn-Gln
(B)Leu-Ala-Lys-Leu-Val
(C)Phe-Arg-Asn-Phe-Gln
(D)Gly-Ala-Lys-Gly-Val
5.Which of the following processes is most similar in prokaryotes and eukaryotes?
(A)alternative splicing of exons
(B)addition of a 3′ poly-A tail to mRNA
(C)addition of a 5′ GTP cap to mRNA
(D)transcription by RNA polymerase
6.The following figure depicts steps in the flow of genetic information in a eukaryotic cell.
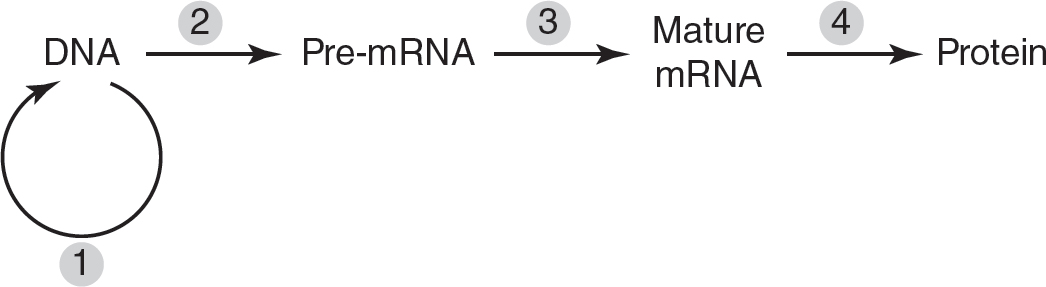
During which step will spliceosomes remove introns?
(A)1
(B)2
(C)3
(D)4
7.The antibiotic tetracycline inhibits bacterial growth by blocking the binding of tRNAs to the ribosome. Which of the following processes is most directly affected by tetracycline?
(A)transcription
(B)exon splicing
(C)initiation of translation
(D)export of proteins from the cell
8.Why do retroviruses have a high mutation rate?
(A)RNA polymerase has a high error rate when reading viral genomes.
(B)DNA polymerase has a high error rate when reading viral genomes.
(C)Viruses use a different genetic code than prokaryotes and eukaryotes use.
(D)Reverse transcriptase has a high error rate.
9.Humans can generate over 1012 different antibody proteins, but humans have fewer than 25,000 genes. Which of the following best explains how this is possible?
(A)Humans acquire new antibody genes when they are infected with pathogens.
(B)Alternative splicing of exons can generate many different transcripts from the same gene.
(C)The error rate in RNA polymerase generates new transcripts for antibody proteins.
(D)Golgi bodies modify RNA transcripts to create new combinations.
10.Which of the following catalyzes the formation of peptide bonds during translation?
(A)RNA polymerase
(B)mRNA
(C)rRNA
(D)tRNA
Short Free-Response
11.Ehlers-Danlos syndrome (EDS) type IV is a result of a mutation in the COL3A1 gene, which results in the deletion of one of the exons in the procollagen transcript.
(a)Describe the location in the cell where the splicing of exons occurs.
(b)Explain the difference between exons and introns.
(c)Predict the effect of the EDS mutation on the structure of the procollagen protein.
(d)Justify your prediction from part (c).
12.A mutation results in the deletion of the TATA box in the promoter of a eukaryotic gene.
(a)Describe the function of the promoter in gene expression.
(b)Explain why promoter sequences are highly conserved in living organisms.
(c)The figure that follows shows the gene and the DNA surrounding it. Draw an “X” on the most likely location of the promoter of the gene.

(d)Explain how the deletion of the TATA box in the promoter would most likely affect the levels of protein produced by the gene.
Long Free-Response
13.The levels of mRNA and protein produced by three different genes (PDHA1, MBP, and INS) were measured in different tissues of the human body. The levels of mRNA and protein expression from these genes were compared to mRNA and protein expression from the gene POLR2A (RNA polymerase II subunit A, which is expressed in all human tissues). The results are shown in the table. “0” indicates that mRNA or protein were not detected in that tissue, and “+,” “++,” and “+++” indicate relatively low, medium, or high levels of mRNA or protein detected, respectively.
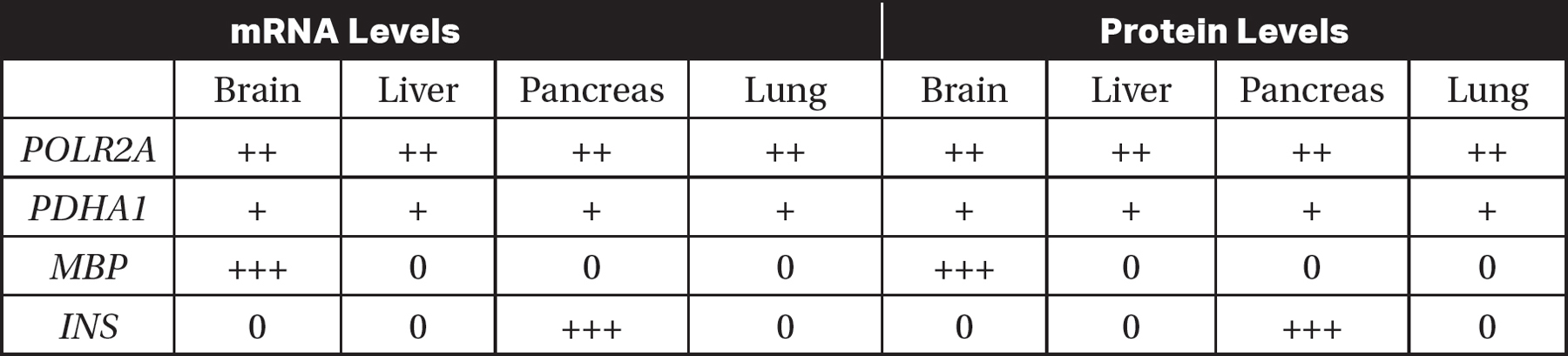
(a)Identify which gene was used as a control in this experiment.
(b)Explain why the gene you selected in part (a) would be an appropriate control for this experiment.
(c)Based on the data in the table, determine which of the genes (PDHA1, MBP, or INS) is most likely involved in glycolysis.
(d)INS is the insulin gene. Insulin is a hormone that is released when blood sugar levels are high. Predict the levels of INS mRNA and protein that would be found in cells in the pancreas when blood sugar levels are low. Justify your prediction.
Answer Explanations
Multiple-Choice
1.(C)Each amino acid is coded by a three base pair codon, so a protein made of 12 amino acids would require a minimum of 12 × 3 = 36 nucleotides. Choice (A) is incorrect because 6 nucleotides would only contain two codons. Since 3 nucleotides are required to make a codon, 12 nucleotides would only be sufficient to code for four amino acids, so choice (B) is incorrect. While 48 nucleotides might code for 12 amino acids if there were introns to be removed, 48 is not the minimum number of nucleotides required, so choice (D) is incorrect.
2.(B)Eukaryotic genes contain introns; prokaryotic genes do not. Prokaryotes do not have the spliceosomes required to remove introns. So if a eukaryotic gene was directly inserted into a bacterial genome, the bacteria would likely try to translate the introns and produce a different protein. Choice (A) is incorrect because prokaryotes and eukaryotes do use the same genetic code. While prokaryotes do not have a nucleus, they do contain RNA polymerase and can perform transcription, so choice (C) is incorrect. Choice (D) is incorrect because rough endoplasmic reticulum is not required for the translation of all proteins.
3.(D)The transcribed mRNA would be antiparallel to the given DNA sequence, so it would start with the 5′ end. Also, in RNA, uracil replaces thymine, and the other base-pairing rules are the same as those found in DNA. Choices (A) and (B) are incorrect because the mRNA is not antiparallel to the DNA sequence. Choices (A) and (C) are incorrect because they contain thymine, which does not appear in RNA.
4.(B)The mRNA transcribed from the DNA sequence in the problem would be 5′ UUA GCA AAG UUA GUU 3′. Using Figure 16.4, the amino acid sequence that would be translated from that mRNA would be Leu-Ala-Lys-Leu-Val.
5.(D)Both prokaryotes and eukaryotes use RNA polymerase for transcription. Only eukaryotes have alternative splicing of exons and the addition of a 3′ poly-A tail and a 5′ GTP cap to mRNA, so choices (A), (B), and (C) are incorrect.
6.(C)Introns are removed from the pre-mRNA to help form the mature mRNA, which is represented by step 3. Step 1 represents DNA replication, so choice (A) is incorrect. Transcription is represented by step 2, so choice (B) is incorrect. Choice (D) is incorrect because step 4 represents translation.
7.(C)If tRNAs could not bind to the ribosome, the initiation of translation could not occur. Choices (A) and (B) are incorrect because transcription and exon splicing do not involve the ribosome, so an antibiotic that interferes with ribosomes would not affect those processes. The export of proteins from the cell involves the Golgi bodies and vesicles, so choice (D) is incorrect.
8.(D)Retroviruses use reverse transcriptase to make a DNA copy of their RNA genome. Reverse transcriptase has a very high error rate, so many mutations occur.
9.(B)Eukaryotes can use alternative splicing of exons to generate multiple mRNA transcripts from one gene, which can lead to the production of multiple proteins from one gene. Humans do not acquire new genes when infected with pathogens, so choice (A) is incorrect. The error rate of RNA polymerase is not responsible for generating new transcripts for antibody proteins, so choice (C) is incorrect. Choice (D) is incorrect because Golgi bodies modify proteins, not RNA transcripts.
10.(C)rRNA catalyzes the formation of peptide bonds during translation. Choice (A) is incorrect because RNA polymerase performs transcription, not translation. mRNA brings the genetic information from the nucleus to the ribosome but does not catalyze the formation of peptide bonds, so choice (B) is incorrect. Choice (D) is incorrect because tRNA serves as an “adapter” molecule that brings the amino acids that correspond to each codon to the ribosome.
Short Free-Response
11.(a)The splicing of exons occurs in the nucleus.
(b)The codons in exons are expressed in the protein and code for amino acids in the protein. Introns are noncoding sequences and do not code for amino acids in the protein.
(c)The EDS mutation would result in a shorter, less functional procollagen protein.
(d)If an exon was deleted, the resulting mRNA transcript would not contain the complete code for the protein. So the translated protein would be shorter and likely less functional.
12.(a)The promoter serves as a binding site for RNA polymerase at which transcription begins.
(b)All living organisms use RNA polymerase, an enzyme that has a consistent three-dimensional shape. All living organisms need similar nucleotide sequences in their promoters that RNA polymerase can recognize.
(c)

(Note that any “X” to the left of the gene is acceptable.)
(d)The deletion of the TATA box in the promoter would most likely make it more difficult for RNA polymerase to recognize the promoter. Less transcription would occur, and less protein would be produced.
Long Free-Response
13.(a)POLR2A was used as the control because the expression of all the other genes was compared to the expression of POLR2A.
(b)POLR2A would be the best gene to use as a control because it is known to be expressed in all human tissues, as stated in the question.
(c)PDHA1 is most likely involved in glycolysis because glycolysis occurs in the cytoplasm of all living cells, and PDHA1 is expressed in all the tissues in the experiment.
(d)Since insulin is released when blood sugar levels are high, it is likely that the levels of insulin would be low when blood sugar levels are low. Therefore, when blood sugar levels are low, it is expected that both the INS mRNA and the insulin protein levels would be low.