Biology Premium, 2024: 5 Practice Tests + Comprehensive Review + Online Practice - Wuerth M. 2023
UNIT 6 Gene Expression and Regulation
18 Biotechnology
Learning Objectives
In this chapter, you will learn:
➜Bacterial Transformation
➜Gel Electrophoresis
➜Polymerase Chain Reaction (PCR)
➜CRISPR-Cas9
Overview
The tools of biotechnology are being used for many purposes: cancer therapies, improvement in agriculture yields, gene therapies for genetic disorders, de-extinction projects (such as the one to bring back the woolly mammoth), and extinction projects (trying to eliminate pathogens that cause human disease), just to name a few. Understanding the basics of how these techniques work, and their possible uses and misuses, is very important. This chapter will briefly review a few basic biotechnology techniques to know for test day.
Bacterial Transformation
Bacterial transformation introduces foreign DNA into bacterial cells. The foreign DNA, usually a small, circular piece of DNA called a plasmid, may integrate into the host cell’s chromosome or remain separate from the host cell DNA in the cell’s cytoplasm. One method of transforming bacteria involves heat shock, in which bacterial cells are mixed with foreign DNA and then quickly exposed to a cold-hot-cold temperature transition. This heat shock creates temporary microscopic pores through which the foreign DNA can enter some of the bacterial cells.
The foreign plasmid DNA needs a selectable marker so that cells that have incorporated the plasmid DNA can be detected. The selectable marker is usually an antibiotic resistance gene that is not present in nontransformed bacterial cells. By growing the transformed bacteria on an agar plate that contains the antibiotic, one can select for the bacteria that absorbed and are now expressing the genes from the plasmid.
If the plasmid contains a gene from another organism, it is called recombinant DNA. Recombinant DNA is simply DNA that has been recombined from different source organisms. DNA can be cut at specific sequences using restriction endonucleases (also known as restriction enzymes), and these pieces of DNA can be recombined and connected with DNA ligases. Some recombinant plasmids may contain the selectable marker as well as a gene that codes for a desired protein product. Bacteria that take in the recombinant plasmid would be capable of producing the product that is coded for by the gene on the plasmid. Many pharmaceutical products (such as insulin) are now produced in large amounts by bacteria that contain recombinant plasmids. Before the advent of biotechnology, the insulin needed for patients with diabetes had to be isolated from the pancreas of animals, which was an expensive process and had the potential of transmitting diseases from other organisms. Now, recombinant insulin made by transformed bacteria is safer and less expensive than insulin was 50 years ago.
Gel Electrophoresis
Gel electrophoresis is a technique that is used to separate DNA fragments by size and by charge. DNA fragments can be created by treating a DNA sample with restriction enzymes, which cut DNA at specific base pair sequences. Due to the redundancy of the genetic code, even organisms with the same protein sequences will have slightly different DNA sequences. Treatment of DNA with restriction enzymes will cut these different DNA sequences at different locations and result in different fragment sizes.
The backbone of the DNA double helix consists of five-carbon deoxyribose sugars and phosphate groups. The phosphate groups have a slight negative charge. DNA samples are loaded into wells at the top of the gel, and an electric current is applied to the gel. A positive cathode is attached to the bottom of the gel and the negative anode to the top of the gel. DNA molecules have an overall negative charge (due to their abundance of the phosphate groups), causing them to migrate toward the bottom (positive end) of the gel.
The gel itself is usually made of agarose and contains microscopic pores through which the DNA fragments migrate. Shorter fragments will be able to travel more quickly through these pores; longer fragments will take more time. This leads to shorter fragments being found at the bottom of the gel, farthest from the well into which the DNA sample was loaded; longer fragments will be closer to the top of the gel. The addition of a DNA marker with fragments of known lengths runs alongside the other DNA samples, providing a “ruler” by which to estimate the size of each fragment produced. Each organism’s pattern of fragments on the gel will be different and can be used to create a unique DNA fingerprint.
Polymerase Chain Reaction (PCR)
Polymerase chain reaction (PCR) is used to amplify specific DNA fragments. PCR can be used to create millions of copies of a specific fragment of DNA. PCR involves cycles of DNA replication using primers that are specific to the beginning and end of the fragment of the DNA sequence that is to be amplified. Each cycle of PCR consists of three stages:
1.Denaturing the DNA—This separates the two strands of the double helix. It is usually done at a high temperature.
2.Annealing of the primers—Once the DNA strands have been separated, the temperature is lowered slightly, and primers that are complementary to the desired DNA sequence are allowed to anneal (form hydrogen bonds with) the beginning and end of the fragment of the DNA sequence that is to be amplified.
3.Extension of the primers—DNA polymerase then adds new nucleotides to the primers, creating two copies of the desired DNA sequence. A heat-stable DNA polymerase is used for this step so that it will not be denatured during the rise in temperature with each cycle of PCR.
Each cycle of PCR doubles the number of copies of the desired DNA sequence. The number of copies of the DNA sequence produced at the end of PCR is equal to 2n, where n is the number of cycles of PCR completed. After just 20 cycles of PCR, over one million copies of the DNA sequence can be generated.
PCR can also be used to rapidly sequence segments of DNA. DNA sequencing is important in forensics, bioinformatics, medicine, and evolutionary biology.
CRISPR-Cas9
The clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 system functions in nature as an adaptive immune system in bacteria. Bacteria can be infected by viruses called bacteriophages. When a bacteriophage infects a bacterium, it cuts up the viral DNA and inserts those pieces between short palindromic repeats in the bacteria’s DNA. This allows the bacteria to store information on which viruses have previously infected its cell. When another virus infects the bacteria, the bacteria compare the DNA from the new virus to these stored DNA sequences. If it finds a match, it cuts the DNA of the virus with the Cas9 enzyme.
While CRISPR-Cas9 is not part of the latest AP Biology Course and Exam Description and likely will not be tested on the AP Biology exam, it is an important and timely topic, so a brief discussion of CRISPR is included here.
CRISPR can be used to edit DNA sequences. The Cas9 enzyme uses a piece of guide RNA to find where to cut DNA. By using synthetic guide RNA pieces that correspond to the desired location of the cut to be made, one can direct the Cas9 enzyme to cut at a specific DNA sequence. If this cut is in the middle of a gene or one of its regulatory regions, this can create a “knockout” of the gene in which the gene is no longer functional. Observing the effects of this knockout can help scientists understand the function of that gene.
Sometimes instead of a knockout, a different DNA sequence at the site cut by the Cas9 enzyme is desired. Inserting multiple copies of a donor DNA sequence into the cell allows the cell’s own DNA repair mechanisms to use the donor DNA as a template. The cell can then replace the DNA sequence at the cut site with a copy of the donor sequence.
Practice Questions
Multiple-Choice
1.Which of the following is used to cut DNA at specific base pair sequences?
(A)DNA ligase
(B)gel electrophoresis
(C)polymerase chain reaction
(D)restriction enzymes
2.A forensic scientist recovers a very small amount of evidence at a crime scene. The scientist would like to amplify the number of copies of DNA in the evidence sample. Which of the following techniques should the scientist use?
(A)bacterial transformation
(B)CRISPR-Cas9
(C)gel electrophoresis
(D)polymerase chain reaction
3.A forensic scientist cuts DNA from a crime scene and DNA from a suspect with the same enzyme. Which tool should the scientist use to separate the DNA fragments according to their size?
(A)bacterial transformation
(B)CRISPR-Cas9
(C)gel electrophoresis
(D)polymerase chain reaction
Questions 4 and 5
A scientist at a pharmaceutical company wants to create bacteria that will synthesize human growth hormone.
4.What should the scientist use to add the DNA code for human growth hormone to a plasmid?
(A)bacterial transformation
(B)DNA ligase
(C)gel electrophoresis
(D)polymerase chain reaction
5.The scientist has successfully created a plasmid that contains the DNA code for the human growth hormone gene. Which technique should the scientist use to insert the plasmid into a cell?
(A)bacterial transformation
(B)CRISPR-Cas9
(C)gel electrophoresis
(D)polymerase chain reaction
Questions 6—8
Huntington’s disease is caused by a short tandem CAG repeat in the HTT gene. Individuals with fewer than 35 CAG repeats in the HTT gene do not develop Huntington’s disease. Individuals with 40 or more CAG repeats will develop Huntington’s disease.
6.Which of the following tools would be most useful in amplifying the number of copies of the HTT gene so that more DNA would be available for analysis?
(A)CRISPR-Cas9
(B)gel electrophoresis
(C)polymerase chain reaction
(D)restriction enzymes
7.Which tool would be most useful for estimating the size of the HTT gene isolated?
(A)DNA ligase
(B)gel electrophoresis
(C)polymerase chain reaction
(D)restriction enzymes
8.A transgenic mouse has the HTT gene. A scientist wants to investigate whether inserting a stop codon into the HTT gene might prevent the mouse from developing the disease. Which of the following tools would be most useful in inserting a stop codon into the HTT gene?
(A)CRISPR-Cas9
(B)gel electrophoresis
(C)polymerase chain reaction
(D)restriction enzymes
9.What property of DNA causes it to move toward the positive electrode in gel electrophoresis?
(A)the nitrogen atoms in the nitrogenous bases
(B)the hydrogen bonds that form between the bases in the two strands of the double helix
(C)the charges on the oxygen atoms in the deoxyribose sugars
(D)the charges on the phosphates in the backbone of the helix
10.Why is a heat-stable DNA polymerase required for the polymerase chain reaction (PCR)?
(A)Heat-stable DNA polymerases add DNA nucleotides at a faster rate than other DNA polymerases.
(B)The high temperatures that are required for each cycle of PCR to separate the DNA strands would denature DNA polymerases that are not heat stable.
(C)Heat-stable DNA polymerases anneal more readily to the PCR primers.
(D)Heat-stable DNA polymerases are more accurate than other DNA polymerases.
Short Free-Response
11.The pAMP-pKAN plasmid contains both the ampicillin resistance gene and a kanamycin resistance gene.
(a)Describe a method for inserting this plasmid into a bacterial cell.
(b)Explain how one could detect which bacterial cells had successfully absorbed the pAMP-pKAN plasmid.
(c)A restriction enzyme makes a cut in the kanamycin resistance gene. Predict the result if a bacterium absorbs this piece of DNA that has been cut by the restriction enzyme.
(d)Justify your prediction from part (c).
12.The comet assay technique is one way of measuring the relative amount of DNA damage in a cell. The nucleus of a cell is placed on a slide coated with an agarose gel. An electric current is applied to the gel, and smaller pieces of DNA will move farther away from the location where the nucleus was placed. These smaller pieces of DNA form the “tail,” and the nucleus forms the “head” of the “comet.” Longer tails indicate greater amounts of DNA damage.
(a)Explain why DNA fragments will move through an agarose gel when an electric current is applied.
(b)List and describe two things that could damage DNA and result in longer tails on the comet assay.
(c)Two cells are treated with a known mutagen of DNA. Cell A has two functional p53 alleles (p53 is involved in DNA repair). Cell B has one functional p53 allele, and its second p53 allele is nonfunctional. On the two templates provided, construct a visual representation of the comet assay results for these two cells.
(d)Explain the visual representations you created on the two templates in part (c).
Long Free-Response
13.A student conducts a bacterial transformation experiment using a strain of E. coli that is not resistant to antibiotics. The student also uses a plasmid (pAMP-pTET) that carries genes for both ampicillin and tetracycline resistance. The four different plates used, and the results of this experiment, are shown in the following figure.
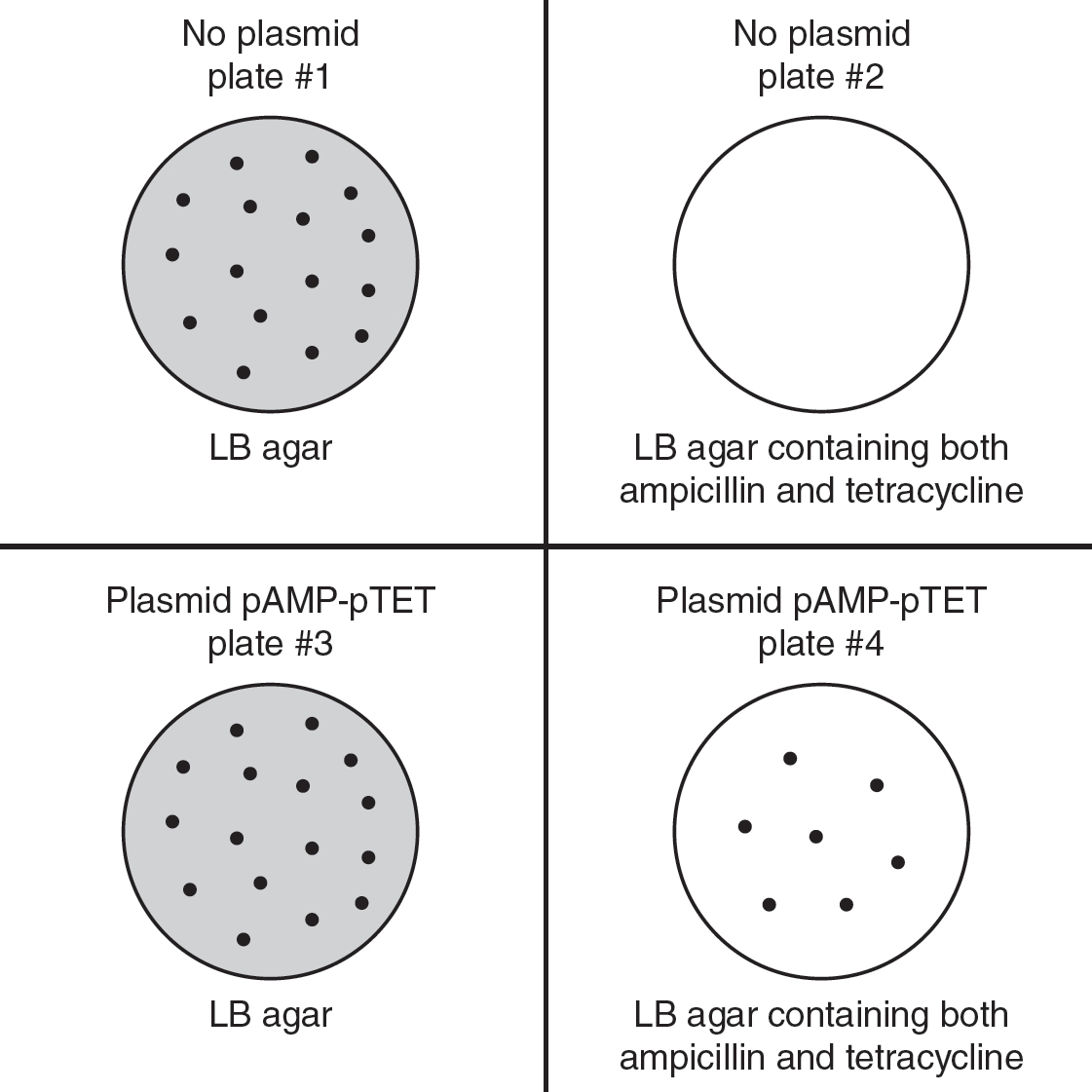
(a)Describe the two enzymes that would be used to create a recombinant plasmid that contained both the ampicillin resistance gene and the tetracycline resistance gene.
(b)Identify the plate that acts as a control to show that the untransformed bacteria were not originally resistant to ampicillin and tetracycline.
(c)Analyze the data, and determine whether the relative numbers of bacteria colonies (seen on agar plate #3 and agar plate #4) are as expected if the bacterial transformation procedure was successful.
(d)Bacteria are removed from plate #4 and are allowed to grow in nutrient medium. Bacteria that contain the F+ (fertility) plasmid can undergo the process of conjugation and transfer some of their DNA to other bacterial cells. Predict the result if bacteria (that were resistant to streptomycin and contained the F+ plasmid) were used to transform the bacteria from plate #4. Justify your prediction.
Answer Explanations
Multiple-Choice
1.(D)Restriction enzymes (also known as restriction endonucleases) cut DNA at specific base pair sequences. DNA ligase connects pieces of DNA, so choice (A) is incorrect. Choice (B) is incorrect because gel electrophoresis is used to separate DNA fragments according to their size. Polymerase chain reaction (PCR) is used to amplify specific sequences of DNA, so choice (C) is incorrect.
2.(D)Polymerase chain reaction is used to make multiple copies of (amplify) specific DNA sequences. Choice (A) is incorrect because bacterial transformation is the process in which bacteria absorb and express foreign DNA. CRISPR-Cas9 is used in gene editing, so choice (B) is incorrect. Gel electrophoresis is used to separate fragments of DNA by size, so choice (C) is incorrect.
3.(C)Gel electrophoresis separates fragments of DNA according to their size. Bacterial transformation involves the uptake and expression of foreign DNA by bacteria, so choice (A) is incorrect. CRISPR-Cas9 is used for gene editing, so choice (B) is incorrect. PCR amplifies specific sequences of DNA, so choice (D) is also incorrect.
4.(B)DNA ligase attaches DNA fragments together, joining them with phosphodiester bonds. Choice (A) is incorrect because bacterial transformation is used to insert naked, foreign DNA into a cell. Gel electrophoresis separates DNA fragments by size, so choice (C) is incorrect. Polymerase chain reaction is used to amplify the number of copies of a specific sequence of DNA, so choice (D) is incorrect.
5.(A)Bacterial transformation inserts naked, foreign DNA into a bacterial cell. CRISPR-Cas9 is used for gene editing, not inserting plasmids into a cell, so choice (B) is incorrect. Choice (C) is incorrect because gel electrophoresis is used to separate DNA fragments according to their size. Polymerase chain reaction is used to amplify the number of copies of a specific DNA sequence, so choice (D) is incorrect.
6.(C)Polymerase chain reaction makes multiple copies of a specific DNA sequence and would be the best choice for amplifying the number of copies of the HTT gene. Choice (A) is incorrect because CRISPR-Cas9 is used for gene editing. Gel electrophoresis separates fragments of DNA by size, so choice (B) is incorrect. Restriction enzymes cut DNA at specific sequences, so choice (D) is incorrect.
7.(B)Gel electrophoresis separates DNA fragments by size and would be most useful for estimating the size of the HTT gene isolated. DNA ligase attaches DNA fragments together, so choice (A) is incorrect. Polymerase chain reaction amplifies specific sequences of DNA, so choice (C) is incorrect. Restriction enzymes cut DNA at specific base pair sequences, so choice (D) is also incorrect.
8.(A)CRISPR-Cas9 is used for gene editing, and with the correct donor DNA, it could be used to insert a stop codon into a gene. Gel electrophoresis separates DNA fragments by size, so choice (B) is incorrect. Choice (C) is incorrect because PCR amplifies the number of copies of a specific DNA sequence. Restriction enzymes cut DNA at specific sequences, so choice (D) is incorrect.
9.(D)The phosphates in the backbone of the DNA double helix have a slight negative charge and are attracted to the positive electrode. The nitrogen atoms in the nitrogenous bases are not charged, so choice (A) is incorrect. Hydrogen bonds between the base pairs do not result in a net negative charge, so choice (B) is incorrect. The oxygen atoms in the deoxyribose sugars are not charged, and thus choice (C) is incorrect.
10.(B)The denaturation stage that occurs during each cycle of PCR requires high temperatures that would denature most enzymes, so a heat-stable DNA polymerase is required for PCR. Heat-stable DNA polymerases do not add DNA nucleotides at a faster rate than other DNA polymerases, so choice (A) is incorrect. PCR primers anneal to the target DNA sequence, not to the DNA polymerase, so choice (C) is incorrect. Choice (D) is incorrect because heat-stable DNA polymerases are not more accurate than other DNA polymerases.
Short Free-Response
11.(a)Bacterial transformation through heat shock could be used to insert this plasmid into the cell. The transition from cold-hot-cold will create temporary small pores through which the plasmids may enter the cell.
(b)Plating the transformed bacteria on an agar plate (that contains both ampicillin and kanamycin) would kill any bacteria that did not absorb the pAMP-pKAN plasmid.
(c)If a bacterium absorbs the piece of DNA that has a cut in the kanamycin resistance gene, the bacterium would still be resistant to ampicillin but not resistant to kanamycin.
(d)The ampicillin resistance gene would still be intact and functional, but the kanamycin resistance gene would not be functional due to the cut made by the restriction enzyme.
12.(a)The phosphates in the sugar-phosphate backbone of DNA have a slight negative charge. They would be attracted to a positively charged electrode.
(b)Any mutagen could damage DNA and result in longer tails. Some examples are ionizing radiation, UV light, and carcinogens (all of which can cause breaks in the DNA sequence).
(c)
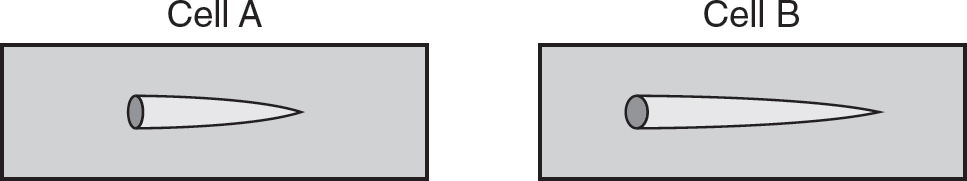
(d)Cell A has two functional p53 alleles and thus would have a greater capacity to repair the DNA damage. Therefore, the tail in the comet assay would be shorter for cell A than the tail for cell B, which only has one functional p53 allele.
Long Free-Response
13.(a)Restriction endonucleases would be used to cut the desired genes from their original plasmids, and DNA ligase would be used to connect them together in a recombinant plasmid.
(b)The plate that acts as this control is plate #2, which contains both ampicillin and tetracycline. This plate shows that the bacteria used in the transformation procedure were not originally resistant to the antibiotics. If no growth occurred on that plate after bacteria that did not receive the plasmid were plated upon it, that would confirm there was no previous antibiotic resistance.
(c)Not every bacterium is expected to absorb the plasmid. So far fewer bacteria colonies would be expected on plate #4 (which contains ampicillin and tetracycline) than on plate #3 (which does not contain any antibiotic). All bacteria, regardless of whether they absorbed the plasmid, would grow on the LB agar in plate #3, while only bacteria that absorbed the plasmid could grow on plate #4. Based on what is shown in the figure, the relative numbers of bacteria colonies on these two plates meets expectations, and thus this bacterial transformation procedure was successful.
(d)The bacteria would now be resistant to three antibiotics: ampicillin, tetracycline, and streptomycin. The bacteria taken from plate #4 would be resistant to ampicillin and tetracycline, and after successful conjugation with the bacteria that have streptomycin resistance, the bacteria would be resistant to streptomycin as well.