THE LIVING WORLD
Unit Four. The Evolution and Diversity of Life
16. Prokaryotes: The First Single-Celled Creatures
16.8. How Bacteriophages Enter Prokaryotic Cells
Bacteriophages are viruses that infect bacteria. They are diverse both structurally and functionally, and are united solely by their occurrence in bacterial hosts. Bacteriophages with double-stranded DNA have played a key role in molecular biology. Many of these bacteriophages are large and complex, with relatively large amounts of DNA and proteins. Some of them have been named as members of a “T” series (T1, T2, and so forth); others have been given different kinds of names. To illustrate the diversity of these viruses, T3 and T7 bacteriophages are icosahedral and have short tails. In contrast, the so-called T-even bacteriophages (T2, T4, and T6) are more complex, as shown by the T4 bacteriophage in figure 16.9a. T-even phages have an icosahedral head that holds the DNA (shown in the cutaway view), a capsid that consists primarily of three proteins, a connecting neck with a collar and “whiskers,” a long tail, and a complex base plate. Many of these structures are also visible in the micrograph of a T4 bacteriophage in figure 16.10a.
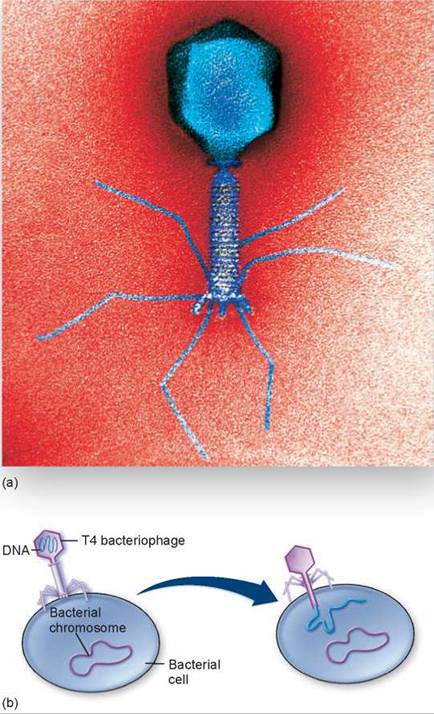
Figure 16.10. A T4 bacteriophage.
(a) Electron micrograph of T4 and (b) diagram of a T4 bacteriophage infecting a bacterial cell.
The Lytic Cycle
During the process of bacterial infection by a T4 bacteriophage, at least one of the tail fibers contacts the lipopolysac- carides of the host bacterial cell wall. The other tail fibers set the bacteriophage perpendicular to the surface of the bacterium and bring the base plate into contact with the cell surface, as seen on the left side in figure 16.10b. After the bacteriophage is in place, the tail contracts, and the tail tube passes through an opening that appears in the base plate, piercing the bacterial cell wall (as shown on the right side of figure 16.10b). The contents of the head, mostly DNA, are then injected into the host cytoplasm.
The T-series bacteriophages and other phages such as lambda (λ) are all virulent viruses, multiplying within infected cells and eventually lysing (rupturing) them. When a virus kills the infected host cell in which it is replicating, the reproductive cycle is referred to as a lytic cycle (see figure 16.11). The viral DNA that is injected in the cell is transcribed and translated by the host cell into viral components that are assembled in the host cell. Eventually, the host cell ruptures and the new lambda phages are released, ready to infect more cells.
The Lysogenic Cycle
Many bacteriophages do not immediately kill the cells they infect, instead integrating their nucleic acid into the genome of the infected host cell (see the lower cycle shown in figure 16.11). While residing there, it is called a prophage. Among the bacteriophages that do this is the lambda phage of Escherichia coli, which is also a lytic phage. We know as much about this bacteriophage as we do about virtually any other biological particle; the complete sequence of its 48,502 bases has been determined. At least 23 proteins are associated with the development and maturation of lambda phage, and many other enzymes are involved in the integration of these viruses into the host genome.
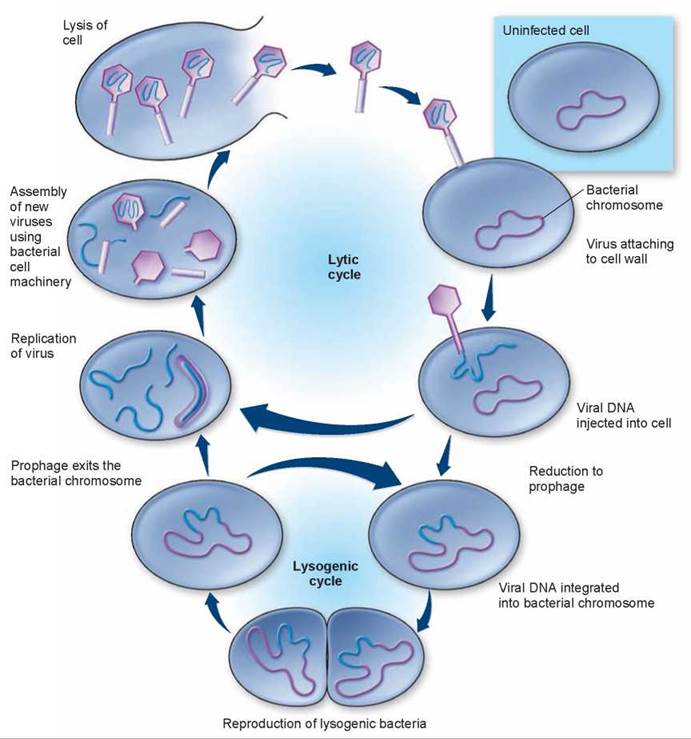
Figure 16.11. Lytic and lysogenic cycles of a bacteriophage.
In the lytic cycle, the bacteriophage exists as viral DNA, free in the bacterial host cell's cytoplasm. The viral DNA directs the production of new viral particles by the host cell until the virus kills the cell by lysis. In the lysogenic cycle, the bacteriophage DNA is integrated into the large, circular DNA molecule of the host bacterium and reproduces. It may continue to replicate and produce lysogenic bacteria or enter the lytic cycle and kill the cell. Bacteriophages are much smaller, relative to their hosts, than illustrated in this diagram.
The integration of a virus into a cellular genome is called lysogeny. At a later time, the prophage may exit the genome and initiate virus replication. This sort of reproductive cycle, involving a period of genome integration, is called a lysogenic cycle. Viruses that become stably integrated within the genome of their host cells are called lysogenic viruses or temperate viruses.
The expression of viral genes integrated into bacterial chromosomes is called gene conversion. An important example of the potential serious effects of viral genes becoming integrated into a bacterial chromosome is provided by the bacterium responsible for the often-fatal human disease cholera. The bacteria Vibrio cholerae usually exist in a harmless form, but a second disease-causing, virulent form also occurs. In this latter form, the bacterium causes the deadly disease cholera. Research now shows that a bacteriophage that infects V. cholerae introduces into the host bacterial cell a gene that codes for the cholera toxin. This gene becomes incorporated into the bacterial DNA, where it is translated along with the other host genes, thereby transforming the benign bacterium to a disease-causing agent.
Lysogenic conversion is also responsible for the presence of toxin genes in (and much of the virulence of) other pathogens like Corynebacterium diphtheriae, which causes diphtheria, Streptococcus pyogenes, which causes scarlet fever, and Clostridium botulinum, which causes botulism.
Key Learning Outcome 16.8. Bacteriophages are a diverse group of viruses that attack bacteria. Some kill their host in a lytic cycle; others integrate into the host's genome, initiating a lysogenic cycle. Bacteriophages transform Vibrio cholerae and other bacteria into disease-causing agents.
Biology and Staying Healthy
Bird and Swine Flu
The influenza virus has been one of the most lethal viruses in human history. Flu viruses are animal RNA viruses containing 11 genes. An individual flu virus resembles a sphere studded with spikes composed of two kinds of protein. Different strains of flu virus, called subtypes, differ in their protein spikes. One of these proteins, hemagglutinin (H), aids the virus in gaining access to the cell interior. The other, neuraminidase (N), helps the daughter virus break free of the host cell once virus replication has been completed. Flu viruses are currently classified into 13 distinct H subtypes and 9 distinct N subtypes, each of which requires a different vaccine to protect against infection. Thus, the virus that caused the Hong Kong flu epidemic of 1968 has type 3 H molecules and type 2 N molecules, and is called H3N2.
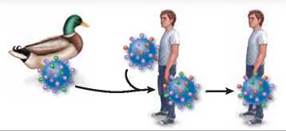
Recombination within humans. A person infected with a flu virus can become infected with another type of flu virus by direct contact with birds. The two viruses can undergo genetic recombination to produce a third type of virus, which can spread from human to human.
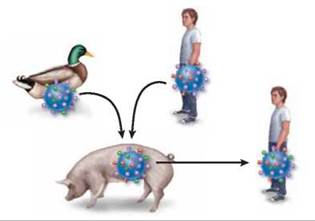
Recombination within pigs. Pigs can contract flu viruses from both birds and humans. The flu viruses can undergo genetic recombination in the pig, to produce a new kind of flu virus, which can spread from pigs to humans.
How New Flu Strains Arise
Worldwide epidemics of the flu in the last century have been caused by shifts in flu virus H-N combinations. The "killer flu” of 1918, H1N1, thought to have passed directly from birds to humans, killed between 40 and 100 million people worldwide. The Asian flu of 1957, H2N2, killed over 100,000 Americans, and the Hong Kong flu of 1968, H3N2, killed 70,000 Americans.
It is no accident that new strains of flu usually originate in the Far East. The most common hosts of influenza virus are ducks, chickens, and pigs, which in Asia often live in close proximity to each other and to humans. Pigs are subject to infection by both bird and human strains of the virus, and individual animals are often simultaneously infected with multiple strains. This creates conditions favoring genetic recombination between strains, as illustrated above, sometimes putting together novel combinations of H and N spikes unrecognizable by human immune defenses specific for the old configuration. The Hong Kong flu, for example, arose from recombination between H3N8 from ducks and H2N2 from humans. The new strain of influenza, in this case H3N2, then passed back to humans, creating an epidemic because the human population had never experienced that H-N combination before.
Conditions for a Pandemic
Not every new strain of influenza creates a worldwide flu epidemic. Three conditions are necessary: (1) The new strain must contain a novel combination of H and N spikes, so that the human population has no significant immunity from infection; (2) the new strain must be able to replicate in humans and cause death—many bird influenza viruses are harmless to people because they cannot multiply in human cells; (3) the new strain must be efficiently transmitted between humans. The H1N1 killer flu of 1918 spread in water droplets exhaled from infected individuals and subsequently inhaled into the lower respiratory tract of nearby people.
The new strain need not be deadly to every infected person in order to produce a pandemic—the H1N1 flu of 1918 had an overall mortality rate of only 2%, and yet killed 40 to 100 million people. Why did so many die? Because so much of the world's population was infected.
Bird Flu
A potentially deadly new strain of flu virus emerged in Hong Kong in 1997, H5N1. Like the 1918 pandemic strain, H5N1 passes to humans directly from infected birds, usually chickens or ducks, and for this reason has been dubbed "bird flu.” Bird flu satisfies the first two conditions of a pandemic: H5N1 is a novel combination of H and N spikes for which humans have little immunity, and the resulting strain is particularly deadly, with a mortality of 59% (much higher than the 2% mortality of the 1918 H1N1 strain). Fortunately, the third condition for a pandemic is not yet met: The H5N1 strain of flu virus does not spread easily from person to person, and the number of human infections remains small.
Swine Flu
A second potentially pandemic form of flu virus, H1N1, emerged in Mexico in 2009, passing to humans from infected pigs. It seems to have arisen by multiple genetic recombination events between humans, birds, and pigs. Like the 1918 H1N1 virus, this flu (dubbed "swine flu”) passes easily from person to person, and within a year it spread around the world. Also like the 1918 virus, the initial wave of swine flu infection triggered only mild symptoms in most people. The 1918 H1N1 virus only became deadly in a second wave of infection, after the virus had better adapted to living in the human body. Like bird flu, public health officials continue to watch swine flu carefully, for fear that a subsequent wave of infection may also become more lethal.