Case Files Biochemistry, 3rd Edition (2015)
SECTION II. Clinical Cases
CASE 5
A 32-year-old man presents to your clinic with complaints of a sore throat. He reports numerous upper respiratory infections during the last 3 months. The patient also states that he required antibiotics for some of these infections. The patient’s sore throat has been present for 4 days and is progressively worsening. He is no longer able to eat solid foods because of the pain. No one else in contact with him has been ill. Patient gives a history of previous intravenous (IV) drug use, but no other significant medical history is given. On examination, the patient is found to have a temperature of 100.0°F (37.8°C) and is in minimal distress from the sore throat. His pharynx is erythematous and has numerous white plaques coating the throat. There is also prominent cervical lymph node enlargement. His chest is clear to auscultation and heart is regular rate and rhythm. A CD4 T-lymphocyte cell count is performed and is less than 200 cells/mm3 (normal > 500 cells/mm3). The responsible organism is composed of the ribonucleic acid (RNA) genome.
![]() What is the most likely diagnosis?
What is the most likely diagnosis?
![]() What is the biochemical mechanism that the pathogen uses to affect the patient’s cells?
What is the biochemical mechanism that the pathogen uses to affect the patient’s cells?
![]() What enzyme is required for this pathogen to affect the host genome?
What enzyme is required for this pathogen to affect the host genome?
ANSWERS TO CASE 5:
HIV
Summary: A 32-year-old man with history of IV drug use has numerous upper respiratory symptoms and adenopathy, and he presents now with sore throat with numerous white plaques. He also has a low CD4 count. The pathogen has an RNA genome.
• Most likely diagnosis: Candida esophagitis secondary to HIV infection.
• Biochemical mechanism of disease: The HIV genome is composed of single-stranded RNA. Reverse transcription of the viral RNA is required to infect the host cells. HIV causes immunosuppression because it has a propensity for attaching to helper T cells (CD4 cells), the cells that support cell mediated immunity.
• Enzyme necessary: Reverse transcriptase.
CLINICAL CORRELATION
This 32-year-old man likely acquired HIV infection by sharing needles with an infected individual. Initially, HIV infection may cause systemic, flulike symptoms, adenopathy, and fatigue. During this phase, the patient experiences viremia. The next phase is largely asymptomatic as the virus slowly causes attrition of the CD4 helper T cells. When the levels of these important cells drop to low levels, the patient cannot fend off organisms commonly colonizing the skin, gastrointestinal tract, or in the air. Treatment for HIV infection includes agents that attack the unique aspects of the virus, such as nucleoside analogue reverse transcriptase inhibitors and protease inhibitors.
APPROACH TO:
HIV
OBJECTIVES
1. Understand normal transcription/translation.
2. Be familiar with reverse transcriptase and mechanism by which HIV works.
3. Know the mechanism of action of HIV medications.
DEFINITIONS
DNA REPLICATION: The process by which DNA is duplicated in the cell takes place during the S-phase of the cell cycle. DNA is duplicated in a semiconservative manner; that is, each new DNA double-strand contains one of the original strands (parent strands) and one of the newly synthesized strands (daughter strands).
HIV: Human immunodeficiency virus; a retrovirus that causes AIDS.
RETROVIRUS: A virus in which the genetic material located in the virus is RNA. The genetic information in the retrovirus must be converted to DNA in the host by the process of reverse transcription.
REVERSE TRANSCRIPTION: The synthesis of DNA from an RNA template.
TRANSCRIPTION: The process by which the information contained in the nuclear DNA is converted to cytosolic messenger RNA (mRNA). This is accomplished by RNA polymerases that synthesize an mRNA that has a complementary sequence to the DNA strand that was used as the template.
TRANSLATION: The synthesis of proteins from mRNA. This is a process that takes place on ribosomes and requires the participation of aminoacyl-charged transfer RNA (tRNA), mRNA, and various initiation and elongation factors.
EXONS: Sequence of nucleotides that appear in mature RNA.
INTRONS: Sequences of nucleotides that do not appear in mature RNA but are excised and do not appear in messenger RNA.
SPLICEOSOME: A complex of small nuclear ribonucleoprotein particles (snRNPs) that catalyze the splicing, or removal of introns, of mRNA precursors.
DISCUSSION
DNA is the biologic blueprint material, which authentically carries all the necessary cellular information and passes it from generation to generation. Thus, it is essential for the cells to preserve the integrity of the DNA and keep it as error free as possible. One is amazed with the versatile DNA molecule, which dictates both the unique and the similar features of the offspring from its parents. What are the processes that take place in a cell to duplicate and interpret this genetic code into functional signals? DNA is duplicated in a semiconservative fashion in a process called DNA replication.
DNA interpretation follows the central dogma (Figure 5-1): first, the DNA is decoded to form mRNA in the nucleus by a process called transcription. Transcription is a complex process involving an enzyme RNA polymerase and several transcription factors. The DNA strand that directs RNA synthesis via complementary base pairing is called the template. RNA synthesis is always unidirectional from 5′ (phosphate) to 3′ (hydroxyl).One can organize transcription in a stepwise fashion:
1. Binding of RNA polymerase to the DNA
2. Formation of the transcription bubble (separation of the DNA strands)
3. Addition of the first RNA residue
4. Addition of the second residue, formation of a phosphodiester bond between the ribonucleic acid residues, and release of pyrophosphate
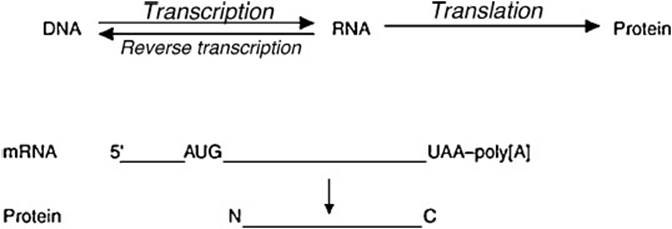
Figure 5-1. The central dogma.
The addition process continues until the termination signal is encountered. Once the nascent RNA chain emerges, it is capped with methylguanylate at the 5′ end and polyadenylated in the 3′ end. This primary RNA transcript is processed further by splicing machinery to yield the mature functional mRNA. Heterogenous nuclear RNA (hnRNA) contains exons and introns. The introns are excised and mature RNA enters the cytoplasm.
The mRNA is then processed in the cytoplasm by a process called translation. Translation involves the ribosomes along with a battery of initiating (IF), elongation (EF), and release (RF) factors. An important rule to remember in translation is that each triplet nucleotide codon codes for a specific amino acid (the 3-letter genetic code). Maintaining the order of amino acids is important to obtain a functional protein. mRNA is translated into protein with the help of tRNA, which brings an amino acid along with it (aminoacyl tRNA) and ribosomes. Ribosomes are in turn composed of numerous proteins and several rRNA molecules. The stepwise process of translation is as follows:
1. The ribosomal unit along with the initiation factors and formyl-methionine (fMet)-tRNA bind close to the initiating codon AUG in the 5′ region of the mRNA.
2. The ribosomal complex consists of 3 sites, namely, A, P, and E. The aminoacyl tRNA binds the triplet codon at the A site. The formyl-methionyl group forms a peptide bond with the amino group of the aminoacyl tRNA in the P site, releasing the now uncharged tRNA fMet, which exits the ribosome assembly through the E site.
3. Elongation continues until the ribosome encounters one of the stop codons UAA, UAG, and UGA.
4. Termination of protein synthesis occurs with the help of release factors that cleave the peptide chain from the tRNA and dissolve the ribosomal complex.
In retroviruses such as HIV in which the genetic material is RNA rather than DNA, reverse transcription occurs. The life cycle of HIV begins when the virion binds to cell surface receptors (CD4 receptors) on helper T lymphocytes. This results in a conformational change that enables the viral coat to fuse with the lymphocyte membrane, thus releasing the viral RNA and viral proteins into the cytosol, including reverse transcriptase and integrase. The RNA genome (Figure 5-2) is reverse transcribed into a double-stranded DNA molecule by utilizing the reverse transcriptase enzyme. This enzyme is an unusual DNA polymerase, because it uses both DNA and RNA as template. It makes DNA from the RNA and uses this in turn to make the second strand of DNA. This double-stranded DNA is transported into the nucleus and is recognized by the viral enzyme integrase, which catalyzes the insertion of the viral DNA into the host genome, thus establishing a permanent infection. The final step is the transcription of the integrated viral DNA producing a large amount of viral RNA, which is packaged in a capsid along with the other essential proteins and can bud from the plasma membrane. Based on the pathogenesis of HIV, it is very important to understand the transcription mechanism of this virus. The HIV life cycle can be simplified and drawn out as shown in Figure 5-3.
![]()
Figure 5-2. Simplified HIV genome.
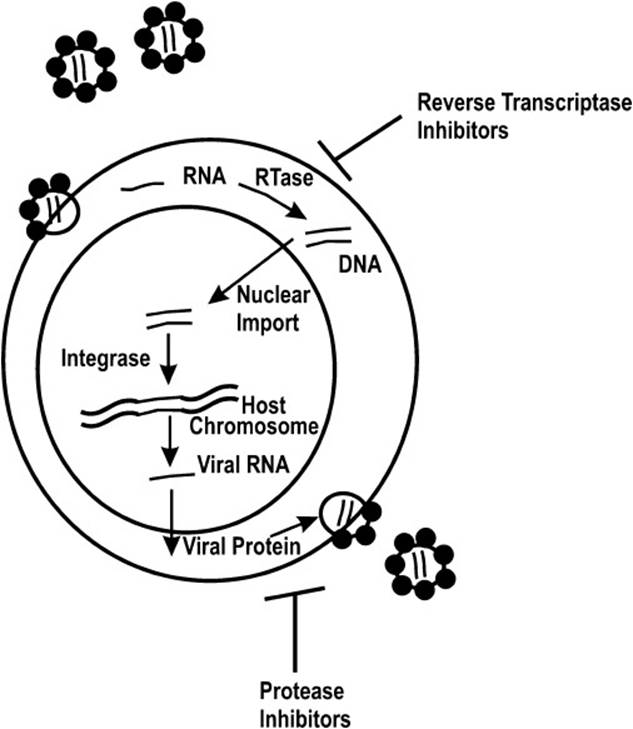
Figure 5-3. Life cycle of HIV.
The long terminal repeat (LTR) contains the enhancer and the promoter regions of HIV. The gag encodes the structural proteins, which help in packaging the RNA of the virus to generate new virus particles. The pol geneencodes both the critical enzymes reverse transcriptase and integrase. The env codes for envelope proteins of the virus that along with the host plasma membrane help the complete virus particle to bud off from the cell. Apart from these proteins, the 3 regulatory proteins, namely, the negative factor Nef, transactivator of transcription Tat, and the regulator of viral gene expression, Rev, can affect viral transcription output. Tat specifically promotes transcriptional activity, while Rev is responsible for switching from early to late HIV gene expression and is located in the 3′ end of the env gene. With the latest research identifying new proteins in addition to the pre-existing knowledge, one wonders what would be the most effective antiretroviral therapy. The available drugs in the market include nucleoside reverse transcription inhibitors and protease inhibitors.
Nucleoside reverse transcriptase inhibitors act as substrates for the reverse transcriptase enzyme and require a phosphorylation event to be activated. The nucleoside drugs lack the 3′-hydroxyl group; therefore, their incorporation into viral DNA will effectively terminate the elongation process. This class includes zidovudine, didanosine, stavudine, lamivudine, and abacavir. As these agents affect very early events in pathogenesis of HIV, they prevent acute infection of susceptible cells but have little effect on already infected cells. The common adverse events of these drugs are lactic acidosis, hepatomegaly, anemia, anorexia, fatigue, nausea, and insomnia. A subcategory to this class of drugs is the nonnucleoside inhibitors, which act by binding close to the active site, inducing a conformation change and thereby inhibiting enzyme function. Some of the members of the class are delavirdine, nevirapine, and efavirenz. A major drawback with these nonnucleoside drugs is that they are effectively metabolized by the cytochrome P450 system and are prone to drug interaction. The most common adverse effects are rash, elevated liver function, and impaired concentration.
Protease inhibitors are targeted toward the HIV proteases, which are essential to activate precursors of gag-pol. HIV protease is essential for infectivity and cleaves the viral polyprotein (gag-pol) into active viral enzymes and structural proteins. The mechanism of action of these drugs is binding reversibly to the active site of HIV protease and blocking subsequent viral maturation. This major class includes saquinavir, ritonavir, indinavir, and nelfinavir. Toxicities of protease inhibitors include nausea, vomiting, and diarrhea.
HIV inhibitor treatment eventually leads to the selection of resistant mutations, one key reason being that the reverse transcriptase is error prone because it lacks the 3′ exonuclease activity. Currently, combinatorial inhibitors with different protein targets; for example, 2 reverse transcriptase nucleoside inhibitors and 1 protease inhibitor, are used in highly active antiretroviral therapy (HAART). To overcome drug resistance and find a definitive cure for HIV infection, several efforts are being made to develop vaccines and also use ribozymes to target HIV mRNA. Because of the molecular understanding of HIV, the future of anti-HIV therapy seems bright and promises an effective cure for patients with AIDS.
COMPREHENSION QUESTIONS
5.1 The current therapeutic strategy for patients who have been infected with HIV is a multidrug regimen known as HAART. One type of drug used in this therapy is a nucleoside/nucleotide analog (eg, didanosine). Which of the following best describes the mechanism of action of these drugs?
A. They inhibit the synthesis of viral proteins.
B. They directly bind to and inhibit reverse transcriptase.
C. They prevent the hydrolysis of the viral polyprotein.
D. They prematurely terminate the DNA synthesized by reverse transcriptase.
E. They inhibit the viral enzyme integrase.
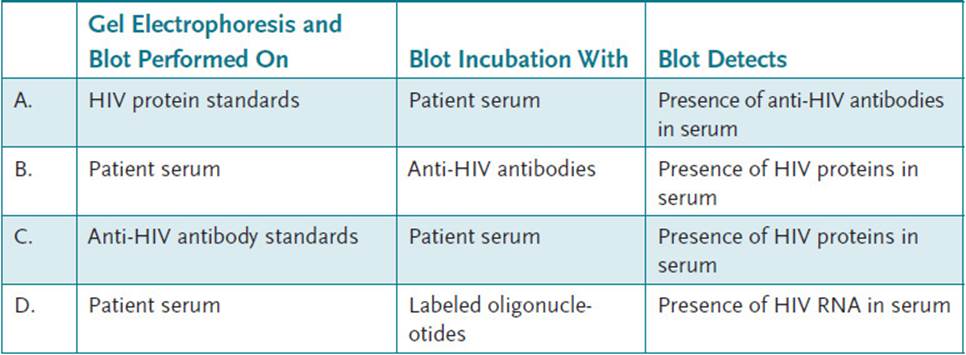
5.2 Enzyme-linked immunosorbent assay is used for routine screening of HIV infection, and the Western blot test has been successfully used as a confirmatory test. Which of the following best describes the strategy used to confirm the presence of an HIV infection by Western blot analysis?
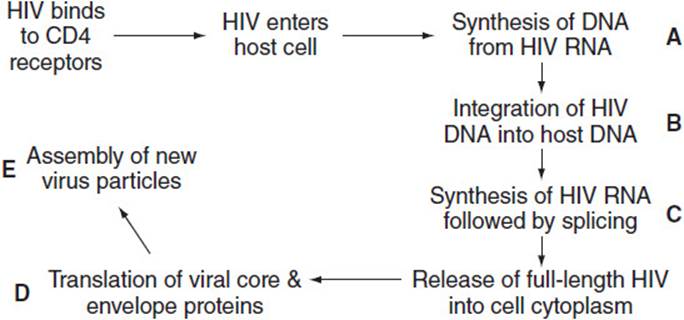
5.3 The following diagram schematically represents the life cycle of HIV. Which of the labeled steps best indicates the site at which an HIV protease inhibitor disrupts the cycle?
5.4 Given the mRNA nucleotide sequence, choose the best protein sequence that will likely result. (Hint: Use the amino acid table in questions 13.2 and 13.3 in Case 13, keeping in mind that [T] and [U] are analogous). mRNA 5′ AUCGGAUGUCUCGGGUUCUGUAAAGGUAAUC 3′
A. Met-Ser-Arg-Val-Leu
B. Ser-Arg-Val-Leu
C. Met-Leu-Ser-Val
D. Ser-Arg-Val-Phe-Phe
E. Pro-Ser-Val-Gly
ANSWERS
5.1 D. The nucleoside/nucleotide analogs like azidothymidine and didanosine are incorporated into the DNA synthesized by HIV reverse transcriptase. Because they do not have a 3′-hydroxyl group, they cannot form a bond with the next nucleotide and the chain is terminated. Host-cell DNA synthesis is not affected because of the nuclear DNA repair mechanisms.
5.2 A. In the Western blot confirmatory test, standard HIV proteins (gag, pol, and env) are separated by gel electrophoresis and then blotted onto a nitrocellulose membrane. The membrane is then incubated with the patient’s serum. Any anti-HIV antibodies will bind to the respective HIV protein on the membrane. Finally, a labeled antihuman antibody is added to indicate the presence of any anti-HIV antibody binding. The test is highly specific; that is, a positive result is highly indicative of HIV infection.
5.3 E. The HIV proteins synthesized by the host cell are produced as a long polyprotein that must be cleaved to the active HIV enzymes and structural proteins. HIV protease inhibitors bind to and inhibit the aspartic protease that hydrolyzes the polyprotein, thus preventing the assembly of infective viral particles.
5.4 A. The initiator codon is AUG and codes for Met; UCU = Ser; CGG = Arg; GUU = Val; CUG = Leu; UAA = stop codon. The mRNA is read in the 5′→ 3′ direction.
BIOCHEMISTRY PEARLS
![]() DNA is decoded to form mRNA in the nucleus by a process called transcription. RNA synthesis is always unidirectional from 5′ (phosphate) to 3′ (hydroxyl), and begins at the 3′ end of the DNA chain toward the 5′ end.
DNA is decoded to form mRNA in the nucleus by a process called transcription. RNA synthesis is always unidirectional from 5′ (phosphate) to 3′ (hydroxyl), and begins at the 3′ end of the DNA chain toward the 5′ end.
![]() The mRNA is processed in the cytoplasm (translation) involving the ribosomes. Each triplet nucleotide codon codes for a specific amino acid (the 3-letter genetic code).
The mRNA is processed in the cytoplasm (translation) involving the ribosomes. Each triplet nucleotide codon codes for a specific amino acid (the 3-letter genetic code).
![]() Messenger RNA is translated into protein with the help of tRNA, which brings an amino acid along with it (aminoacyl tRNA) and ribosomes.
Messenger RNA is translated into protein with the help of tRNA, which brings an amino acid along with it (aminoacyl tRNA) and ribosomes.
![]() Some viruses, such as HIV, have RNA genomes, and they usually are reverse transcribed into a double-stranded DNA molecule by utilizing the reverse-transcriptase enzyme.
Some viruses, such as HIV, have RNA genomes, and they usually are reverse transcribed into a double-stranded DNA molecule by utilizing the reverse-transcriptase enzyme.
REFERENCES
Levy JA. HIV and the Pathogenesis of AIDS. 2nd ed. Washington DC: ASM Press;1998.
Lodish H, Berk A, Kaiser CA, et al. Molecular Cell Biology. 7th ed. New York: Freeman; 2012.
Raffanti S, Haas DW. Anti-bacterial agents. In: Brunton L, Chabner B, Bjorn Knollman, eds. Goodman and Gilman’s The Pharmacological Basis of Therapeutics. 12th ed. New York: McGraw-Hill; 2010.