CHEMICAL BIOLOGY
Peptide Combinatorial Libraries
RuiWU Liu, University of California Davis, Sacramento, California Kit S. Lam, University of California Davis, Sacramento, California
doi: 10.1002/9780470048672.wecb434
Peptide libraries, which consist of a few thousands to tens of millions of distinct peptides, can be generated combinatorially through biologic or synthetic approaches. The phage-display peptide library method is simple and economical, and peptides with various natural protein folds can be generated by this approach; but this biologic method generally is limited to peptides that contain only L-amino acids. Synthetic combinatorial libraries, however, can accommodate D-amino acids, unnatural amino acids, and even organic moieties, which makes these approaches highly versatile. These synthetic methods include the spatially addressable parallel library method, the combinatorial library method that requires deconvolution, the one-bead one-compound (OBOC) combinatorial library method, the self-assembled peptide nucleic acid (PNA) encoded chemical microarrays, and the synthetic library method that requires chromatography selection. Various methods to screen these combinatorial libraries have been developed. For example, phage-display peptide libraries can be screened for the ability of phages to bind to unique immobilized target proteins or whole cells, peptide microarrays can be screened for their ability to interact with fluorescent proteins, combinatorial library methods that require deconvolution can be screened by standard solution-phase assays, and OBOC libraries can be screened by on-bead binding or functional assays or several solution-phase cell-based or biochemical assays. Combinatorial peptide library methods are enabling technologies that have proven to be very useful in basic research and drug discovery.
A combinatorial peptide library is a collection of a few thousands to hundreds of millions of different peptides. Geysen et al. (1) opened the door of the combinatorial library when they reported the first combinatorial peptide library (96 peptides) that was generated with the multipin technology in 1984. Since then, several different combinatorial approaches have been developed to generate libraries that contain many peptides with discrete sequences from which bioactive peptides can be isolated and/or identified. In the early reports in this field, synthetic peptide libraries were generated and screened against antibody molecules or streptavidin (1-4). About the same time, the phage-display peptide library method was created (5-7). Since 1990, several totally different combinatorial library methods have been introduced. These methods enable one to generate and screen easily and rapidly libraries with millions to billions of random peptides. The screening of combinatorial peptide libraries allows for the rapid discovery of peptide ligands or substrates against a variety of biologic targets. The principle for selecting new ligands by means of combinatorial peptide libraries exploits the natural diversity of protein-protein and protein-ligand interactions. It has emerged as a powerful tool in the study of many biologic systems. The amino acid sequence of an “active” peptide then is determined. Biologic peptide libraries, such as those that involve filamentous phage-display peptides, typically can accommodate only the 20 natural amino acids. In contrast, synthetic peptide libraries have the potential to incorporate D-amino acids and other unnatural amino acids, as well as specific secondary structures or scaffolding structures that may enhance biologic activity. In addition to amino acids, biologic building blocks such as monosaccharides, nucleotides, lipids, or even small organic moieties can be used. The advent of these peptide library methods not only facilitates the drug discovery process but also provides important information for the fundamental understanding of molecular recognition. In this mini-review, we shall focus our discussion on the preparation, screening, structure elucidation, and application of various peptide library methods.
Peptide Library Methods
In essence only two approaches exist to generate combinatorial peptide libraries: biologic and synthetic library approaches. According to the different techniques used, the synthetic library approach can be divided additionally into five methods: 1) the spatially addressable parallel library method (1, 4, 8), 2) the combinatorial library method that requires deconvolution (3, 9), 3) the one-bead one-compound (OBOC) combinatorial library method (2, 10), 4) the self-assembled peptide nucleic acid (PNA) encoded chemical microarrays (11), and 5) the synthetic library method that requires chromatography selection (12). Although the focus of this review is on peptide libraries, it is important to point out that the five synthetic methods can be applied readily to the generation of small molecule, peptidomimetic, and macrocyclic libraries.
Each peptide library method has its own advantages and disadvantages. A comparison of these methods with respect to cost, resources, and uses is shown in Table 1.
Table 1. Comparison of selected combinatorial library methods
|
Combinatorial library methods |
Advantages |
Disadvantages |
|
|
Biologic library methods |
Phage-display |
Provides up to 109 peptide entities; longer peptides (e.g., 30 amino acids) with tertiary folds possible; molecular biology and DNA sequencing technique available in many laboratories; inexpensive; available commercially and from many research laboratories; and library can be expanded, aliquoted, and stored frozen. |
Limited to peptide libraries with eukaryotic amino acids; limited to binding and simple functional assay. |
|
Spatially addressable parallel library method |
Multi-pin technology |
Structure determination not needed; pin library can be screened directly for binding; releasable peptides from higher loading lanterns can be screened by multiple-solution phase assays; moderately expensive pins and crowns can be used a few times without obvious loss of activity; and commercially available. |
Peptide library is relatively small; pin library with linker effect; and limited to binding and simple functional assay. |
|
|
SPOT synthesis |
Structure determination not needed; convenient on-paper assay; moderately expensive; spot synthesis equipment and custom-spot peptide membrane are commercially available. |
Peptide library is relatively small; limited to binding and simple functional assay if bound peptides are used for the assay; releasable peptides possible, but each in small quantity; and peptide spot membrane generally not recyclable for subsequent use. |
|
|
Nanokan technology |
Easy readout with radiofrequency tags; applicable to standard solution-phase assay; solution peptides can be used for multiple assays; and commercially available. |
Peptide library is relatively small; inefficient unless split-mix synthesis used; peptide requires cleavage from resin for subsequent solution-phase screening; and equipment and supplies are very expensive. |
|
|
|||
|
|
The 96-deep well plate system |
Applicable to standard solution-phase assay; solution peptides can be used for multiple assays; and robotics for automated synthesis are commercially available. |
Inefficient synthesis unless fully automatic; equipment and supplies are expensive. |
|
|
Peptide microarray |
Microassay possible, saving expensive and precious assay reagents; moderately expensive; replicates of peptide chips can be made; and peptide chips are available commercially or can be custom-made. |
Linker effect; in situ synthesis not available widely; spotting technique is rapid but requires synthesis of individual compound separately; limited to on-chip binding and some functional assays; and peptide chip generally not recyclable for subsequent use. |
|
Synthetic library methods that require deconvolution |
Iterative approach |
Many peptides can be synthesized and analyzed rapidly; can be applied to standard solution-phase assay; inexpensive; and can be synthesized easily by experienced peptide chemist. |
Peptide mixture; requires multiple separate iterative synthesis; not as efficient as the positional scanning method (below); and library not commercially available. |
|
|
Positional scanning |
Same as iterative approach (above) except it is more efficient and less synthesis is required; libraries can be aliquoted and used for multiple assays. |
Peptide mixture; results may be ambiguous if multiple hits with different motifs are present in the mixture; and library not commercially available. |
|
One-bead one-compound library method(OBOC) |
On-bead screening |
Highly efficient synthesis and screening; each peptide is spatially separable, therefore multiple different motifs can be identified; applicable to both binding and functional assays; inexpensive; and can be synthesized easily by experienced peptide chemist. |
Linker effect unpredictable until tested; chemical structure of positive beads has to be analyzed; and library not commercially available. |
|
|
Releasable assay |
Highly efficient synthesis; screening applicable to both binding and functional assay. |
Releasable chemistry more involved but is still much more efficient than many other library techniques; new solid support that easily can release compounds needs to be developed; and library can not be reused and is not commercially available. |
|
Self-assembled PNA encoded chemical microarrays |
|
Library decoding on DNA chip is highly efficient; mix-split synthesis possible. |
Synthesis of PNA tag is cumbersome; requires special DNA chip for decoding; limited to binding and simple functional assay; and not commercially available. |
|
Synthetic library method that uses affinity chromatography selection |
|
Library synthesis is easy; inexpensive; and can be synthesized easily by experienced peptide chemist. |
High background because of nonspecific binding; only predominant motif can be identified; limited application; and library not commercially available. |
|
|
|||
Biologic Library Method
The first biologic peptide library was reported by Parmley and Smith in 1988 (13). They developed the “fusion phage” by inserting foreign DNA fragments into the encoding gene of the pIII protein. This development led to the expression of an L-amino acid-containing peptide on the virion surface that did not affect virus infectivity. Each phage particle has five copies of the peptide on one end. The M13 phage is the most widely used phage-display system because of its high capacity for replication and its ability to receive large DNA inserts into its genome. Trinucleotide sequences that encode specific amino acids can be constructed and inserted into the phage genome either to increase diversity or to introduce a bias into the clone population. This very powerful method enables the researcher to routinely generate 108—109 different phages. Such a library can be panned against a target molecule by standard protocols. Other biologic peptide libraries include plasmid libraries, polysome libraries, fimbria libraries, yeast-display libraries, bacterial flagella libraries, mRNA-display libraries, ribosome-display libraries, and other more recently developed in vitro selection methods. For a review on biologic libraries please see Reference 14.
Biologic library methods enjoy the following advantages over synthetic peptide library methods: 1) Many peptide entities can be generated easily and economically, 2) the biologic library method can take advantage of known protein folds (e.g., immunoglobulin fold, zinc-finger fold, or conotoxin fold) by grafting random oligopeptides onto such tertiary folds, and 3) less restriction exists on the size of the peptide (e.g., 30 amino acids) one can generate. However, the biologic approaches suffer from three major disadvantages: 1) With the current systems, only the natural L-amino acids (20 eukaryotic amino acids) can be incorporated into these libraries, and incorporation of unnatural amino acids or other organic subunits into these libraries generally is not feasible, 2) although simple disulfide cyclization is feasible and longer peptides with specific protein folds (such as immunoglobulin folds) may be used, complicated bicyclic, compact scaffolding, branched structures, or molecules with a special chemistry of cyclization are impossible, and 3) the screening assays of the biologic libraries generally are limited to the binding assays (e.g., panning) and some functional assays such as protease substrate determination (see below). These methods generally are not applicable to most standard drug screens that use solution-phase assays. Synthetic peptide libraries, however, can overcome many of these limitations.
Synthetic Library Methods
Spatially addressable parallel library method
Peptide libraries can be synthesized on solid-phase support by using spatially addressable high-throughput synthetic methods (manual or automated). The amino acid sequence of each of these peptides is predetermined. The reported techniques based on this strategy include multipin technology, SPOT synthesis, NanoKan, peptide microarray, multisyringe system, and the 96 deep-well plate system. These methods are labor-intensive but can be facilitated by robotics. Depending on the library method used, the peptide library is screened either by a direct solid-phase binding assay or by a solution-phase assay. The chemical structure of the positive peptide then is decoded by location. The major drawback of this method is that only limited number of peptides can be synthesized and therefore the library generally is rather small.
Multipin technology
As mentioned earlier, the first peptide library with a limited number of peptides was generated by using multipin technology in 1984 (1). In this method, acrylic acid is irradiation-grafted onto the tip of polyethylene pins. Each pin is inserted into an adapter that fits over a 96-well polypropylene plate with a standard microtiter plate footprint. Each well of the microtiter plate then serves as a separate reaction vessel for the amino acid-coupling steps. In the original experiments, ~50 nmol (ideally) of a single peptide was linked covalently to the spherical head of each pin. Today, the multipin system is available commercially from Mimotopes (San Diego, CA) as Pepsets. The peptide loading of each pin has been increased significantly by introducing the so-called “SynPhase™ Lantern” (e.g., 75 μmol for the SynPhase PS A-Series Lanterns) or “SynPhase™ Crown” (e.g., 8.3 μmol for the I-Series Crown) to fit into the tip of each pin.
Tea bag and nanokan technique
In 1985, Houghten (15) introduced the “tea bag” method for simultaneous multipeptide synthesis. In this method, peptide synthesis occurs on resin beads that are sealed inside labeled, porous polypropylene packets. At the end of the synthesis, the individual peptides are liberated from the resin. In 1995, this method was improved by using minibaskets (e.g., Nanokan) to encapsulate radio-frequency microchips (Rf tagging) and a sample of resin beads for peptide synthesis (16, 17). Each of these minibaskets can be scanned with an electronic reader before or right after each coupling cycle during the “split-mix” synthesis (see Fig. 1a) (2, 3, 18). Therefore, the synthetic history of each minibasket can be traced. At the end of the synthesis, compounds can be cleaved off the resin beads inside each minibasket and placed in a 96-well plate to form a spatially addressable compound library. The main advantage of this method is that it offers considerable synthetic flexibility as one may use any resin bead in the synthesis. A few thousand to ten thousand individual peptides can be generated easily. Furthermore, peptides can be produced in sufficient quantity (500 μmol) for purification and complete characterization if desired. The IRORI Nanokan® system is available commercially from Discovery Partners International (San Diego, CA). However, the IRORI system is very expensive. A less expensive encoding system, called Encore (Torviq Inc., Granger, IN), uses “necklace” color tags and multicolor reaction vessels to encode the synthetic history. This system also uses the “split-mix” synthesis strategy to generate compound libraries with a limited number of compounds.

Figure 1. The ''split-mix synthesis'' method to generate a one-bead one-compound combinatorial library (a) and a number of permutations for random peptide libraries (b). P, E, and T are building blocks (in this case amino acids).
SPOT synthesis technique
A peptide library can be synthesized using the SPOT synthesis technique to form a low-density peptide spot array (e.g., 25 spots/cm2). In this method, different peptides are synthesized in situ as low-density arrays on cellulose membrane or paper (8). The volume of Fmoc-amino acids and coupling reagents dispensed creates a specific SPOT size that determines both the scale of reaction and the absolute number of peptides that can be arranged on an area of a membrane. Cotton (another form of cellulose) and polystyrene-grafted polyethylene film segments also have been used as solid supports. Recently, polymeric membranes that are chemically, mechanically, and thermally more stable have been developed, which include hydroxy-functionalized PEG acrylate polypropylene membranes and an amino-functionalized ester-free PEG methacrylamide polypropylene membrane. SPOT synthesis is amenable to miniaturization and automation. Automatic instruments such as the Auto-SPOT Robot ASP222 (Intavis Inc., Cologne, Germany) for SPOT synthesis are available commercially. The advantages of the method are that it is simple, not too expensive, and capable of providing sufficient quantities of peptides for various applications.
Peptide microarrays
Peptide microarrays are prepared by immobilizing many peptide molecules on the surface of a solid support in a small area in an addressable fashion. The immobilization can be achieved via in situ synthesis or chemical ligation through a covalent bond. A hydrophilic linker between the solid surface and the peptide usually is added to minimize steric hinderance caused by the solid support. The most commonly used solid support for microarray printing is a standard microscope glass slide. Other solid supports also have been used such as polystyrene, nitrocellulose membranes, PVDF membranes, Hybond ECL membranes, gold surfaces, and chemical vapor deposited diamond films.
Peptide arrays on paper or cellulose membranes generated by SPOT synthesis generally are low density, even with the commercially available automatic SPOT synthesizer. Such a low-density peptide chip (several thousand peptides) now is available commercially, for example, PepChip™ microarray from Mimotopes. Foder et al. (4) first described the high-density peptide microarray using the photolithographic light-directed parallel synthesis method. The disadvantage of this method is that it requires building blocks (in this case, amino acids) protected with photolabile protecting groups (e.g., 6-nitroveratryloxycarbonyl) that are not available yet commercially. To address this problem, Gao et al. (19, 20) developed digital photochemistry for parallel synthesis of peptides to prepare a peptide microarray. In this method, they combined light-directed synthesis (controlled by computer) with microfluidics so that photo-generated acid (e.g., H+SbF6-) can be generated in situ (with light) to remove the protecting groups of standard commercially available building blocks such as Boc-protected amino acids. More recently, the same group has developed a novel photogenerated base (PGB) applicable to Fmoc-chemistry for the parallel synthesis of a peptide on a microarray. When fully optimized, this versatile in situ, high-throughput parallel synthesis of peptide microarrays would offer unprecedented opportunities for creating various high-throughput detecting and sensing devices, which would enable a broad range of biochemical and biomedical applications (21).
We reported site-specific ligation of peptides to a carrier protein or polymer before printing onto the glass slide with an automatic arrayer to form a peptide microarray (22). Recently, Dikmans et al. (23) reported a novel process for manufacturing multipurpose high-density peptide microarrays termed “Spotting compound-support conjugates” (SC2). In this method, a trifluoroacetic acid (TFA) cocktail containing > 80% TFA plus scavengers is used to solubilize the peptide-cellulose of each SPOT. The peptide-support conjugates then are precipitated, redissolved in dimethyl sulfoxide (DMSO), and then printed on glass slides.
Synthetic Library Methods that Require Deconvolution
The term “deconvolution” has been used to describe the process whereby the active molecule or molecules in a library are identified, usually by the iterative testing of mixtures of compounds for a specific biologic property. Using the results from biologic assays, the identity of the active component may be deduced without the need to determine directly or indirectly its chemical structure. The two main deconvolution approaches are the iterative process (24) and the positional scanning (25). In the iterative approach first reported by Geysen et al. (24) in the multipin system, a progressive selection is performed by choosing one amino acid at a time for each position. Sublibraries are generated based on the result of the previous one; therefore, the sequence is obtained step by step. In the positional scanning method reported by Pinilla et al., sublibraries of peptides with an amino acid fixed at one designated position but randomized in other positions are prepared and tested for biologic activities. Based on the biologic assay results, the amino acid sequence of the active peptide can be deduced at the end of the process. This method assumes that the contribution of each amino acid residue to the biologic activity is independent of each other. It works very well only if one predominant motif exists for the target protein. An interpretation of results from targets with multiple binding motifs likely will be difficult.
OBOC Library Method
In 1991, we first reported the OBOC concept to synthesize peptide libraries using a “split-mix synthesis” method (Fig. 1a) (2). The peptide is synthesized on resin beads such as a 90 pm diameter TentaGel resin (Rapp Polymere, Tubingen, Germany). Because each bead is in contact with only one amino acid at a time during each coupling cycle and the reaction is driven to completion, each bead expresses a single peptide entity (Fig. 1a) and carries about 100 pmol peptide. Because the peptide beads in an OBOC combinatorial library are spatially separable, an OBOC library can be considered as a huge chemical microarray that is not addressable.
The main advantages of the OBOC method include: 1) A large number (e.g., 106—108) of peptides can be synthesized (Fig. 1b) and screened rapidly and simultaneously by using either one or a combination of both on-bead and solution-phase assays, and 2) multiple peptide ligands with completely different motifs often can be identified in a single screening. This occurrence is the opposite of the (convergent) iterative approach (see above) in which multistep synthesis and screening result in the emergence of only one, but not necessarily the best, solution motif.
One major disadvantage of the OBOC method using the on-bead screening method is that each library compound is tethered to the solid support via a linker such as polyethylene glycol and may result in steric hindrance between the cellular receptor and the library compound. However, in some instances the linker may be beneficial, for example, the linker can be used as a convenient handle to link the cancer-targeting ligand to the therapeutic payload.
Self-Assembled PNA-Encoded Chemical Microarrays
In this method, peptides or small molecules are prepared by the “split-mix synthesis” method and cleaved from the resin to form an encoded solution-phase library such that each library compound is tethered to a PNA code via a hydrophilic linker (11). The library then is mixed with the target protein and later exposed to planar oligonucleotide microarrays of predetermined sequences. Alternatively, the encoded soluble library can be hybridized to the oligonucleotide microarrays before incubation with the target protein.
Synthetic Library Method that Requires Chromatography Selection
In this method, the peptide library is an equimolar mixture of random peptides in solution phase from which ligands can be isolated with affinity chromatography (12). It usually is synthesized on solid support with a “split-mix synthesis” method. The peptides then are cleaved from the resins, and the solution-phase peptide library is loaded onto an affinity column with an immobilized receptor. After thorough washing, the bound peptides are eluted and microsequenced. Major concerns about this method include nonspecific binding and uninterpretable results if more than one predominant motif is present in the mixture. Additionally, enough purified peptide must be retrieved for accurate microsequencing or mass spectrometry analysis. In general, because of the high background from nonspecific binding, the affinity selection method, with rare exceptions, can be applied only to a relatively small peptide library (e.g., <10,000 peptides).
Peptide Library Screening
Peptide library screening can be divided into two categories: solid-phase screening, in which peptides still are tethered to the solid-phase support, and solution-phase screening in which the peptides are released from the solid support and tested in solution. Solid-phase screening is generally easier with higher throughput than is solution-phase assay. However, the linker that tethers the peptide to the solid support may interfere with the interaction between the peptide and its receptor. In addition, the multivalent binding between the immobilized synthetic and phage-display peptide gives little information about the real binding affinity between the receptor and an univalent soluble peptide. Different assays such as ELISA, cell-based cyctotoxic assay, antimicrobial assay, affinity chromatography, and radiometric and fluorescence-based assays can be used to screen peptide libraries. Surface plasmon resonance spectroscopy (e.g., Biacore) is a powerful technique to screen soluble and unlabeled peptides, which allows the real-time measurement of soluble peptide binding to different targets of interest immobilized on a chip such as proteins, sugars, fatty acids, nucleic acids, cell membranes, and even whole cells. However, commercially available equipment (e.g., Biacore) that uses label-free detection equipment is generally low throughput. In the last few years, several reports have applied label-free optical detection methods to screen peptide or chemical microarrays (26, 27).
To screen large libraries optimally, carefully selected high- throughput assays should be employed. The choice of the assay system used largely depends on the combinatorial library method used to construct the library, the availability of reagents such as enzymes, antibodies, and radiolabeled ligands, and of course the biologic target itself. To isolate the few active peptides from the library successfully, it is essential to have a robust and accurate screening method that can screen large libraries rapidly. One may use several assay systems serially first to narrow down the potential ligands and then retest them for biologic effect and crossreaction with other unwanted targets.
Phage-Display Library Screening
The phage-display peptide library can be screened easily by panning (e.g., binding to an acceptor-coated petri dish). The phages that bind specifically to an acceptor molecule will be isolated and enriched via several cycles of panning and amplification in Escherichia coli (E. coli). By the end of the panning, the bound phagemids are eluted, cloned, and processed for subsequent identification via DNA sequencing. The phage-display peptide library also has been screened with intact cells in cell culture to identify ligands that bind cell surface receptors. Peptides with a propensity to bind and then enter intact cells can be selected by eluting the phagemids off the cell surface before lysing the cell for phage recovery. Ligands identified through in vitro protein or cell panning need to be validated and evaluated for in vivo targeting. Phage-display peptide libraries also have been screened in vivo in experimental animal models and humans. For example, different libraries have been screened for binding to the vasculature of various organs and tumors by injecting the phage library intravenously into live animals. Organ- or tumor-bound phage then are amplified in E. coli and screened a second and third time before sequence analysis. In a recent study by Krag et al. (28), patients with late-stage melanoma, breast, and pancreatic cancer were infused with random phage-display peptide libraries or phage-display short chain Fv antibodies in either single or multiple panning experiments. For more information about using the phage-display peptide library approach to identify cancer-targeting ligands, please see our recent review (29).
In some cases, functional assays have been used to screen phage-display peptide libraries. For example, a phage-display hexapeptide library was constructed with an epitope tag distal (N-terminus) to screen for peptide substrates of a specific protease. All the phages were captured by an immobilized anti-epitope antibody. After incubation with a tissue plasminogen activator (tPA), phages that expressed a peptide substrate for tPA were released for subsequent rounds of selection. A similar approach was applied to discover peptide substrates for HIV-1 protease.
Spatially Addressable Parallel Library Screening
The spatially addressable parallel libraries can be screened using either a solid- or a solution-phase assay depending on the method used to generate the library. In solid-phase assays, the peptide still is linked covalently to the solid support, such as a pin, SPOT-membrane, or glass surface. The molecular target can be a purified protein, a protein mixture, an enzyme, an intact cell, or a whole organism such as bacteria or virus. For example, enzyme-linked immunosorbant assay (ELISA) assay has been used for a multipin or spot-synthesis peptide library. Binding assays can be used to determine binding specificities, such as for the identification of cell-specific surface markers. One unique feature of chemical microarrays for cell adhesion analysis is that in addition to obtaining binding profiles of different cells to many ligands, one also can examine the signaling response of each binding event. Such signaling profiles can be determined with many fluorescent-labeled antibodies in conjunction with confocal microscopy. Functional properties also could be examined by using other assays to measure specific biologic effects that the ligands have on the target.
In the solution-phase assay, the peptides are cleaved from the solid support, which enables the soluble ligands to interact freely with the target molecule or cell. The peptides are put into 96-, 384-, or 1536-well plates for high-throughput biologic analysis with the aid of robotics. Many conventional assays can be applied to the screening including cell-based cytotoxic assays such as MTT and XTT assays, antimicrobial assays, competitive ELISA assays, radioimmunoassays, radioligand binding assays, fluorescent polarization assays, time-resolved fluorescence assays, fluorescent protein-based recombinant cell bioassays, scintillation proximity assays, and cell-based calcium flux assays.
OBOC Peptide Library Screening
OBOC peptide library can be screened on bead or in solution phase if the bound peptides are released via a cleaveable linker. The on-bead assays include binding and functional assays. In the binding assays, the target of interest could be a purified protein, a protein complex, cell lysates, live cells, or a whole microorganism. In the case of protein screening, protein first is incubated with a library of immobilized ligands. Protein-ligand interaction can be visualized with an appropriate reporting group such as a biotin, an enzyme, a fluorescent probe, a color dye, or a radionucleotide. The biotinylated protein can be detected with a colorimetric assay by using a streptavidin-alkaline phosphatase conjugate followed by color development with a 5-bromo-4-chloro-3-indoyl phosphate. Fluorescent microscopy has been used successfully to screen bead libraries with fluorescent probes. An organic fluorescent dye or quantum dots have been used as a fluorophor to label the target protein. Radiolabeled probe is useful but more tedious than the colorimetric method.
The use of intact live cells or whole microorganisms to screen OBOC peptide libraries is very attractive because purified target proteins are not needed in such a screening. For microorganisms such as a virus or a bacteria, a reporter antibody or a specific dye that stains such microorganisms may be useful. For intact cells, reporter probes are not needed because cells that bind to a bead can be visualized easily under a dissecting microscope. The positive beads can be washed off the cells and sent for sequencing. To enhance the quality of the results more, the positive beads can be retested against either the same cells or different cells lines before being sequenced to ensure that they are true positives. Alternatively, one may label one cell type with calcein and leave the negative control cell line unlabeled. Beads that bind only to fluorescent green cells but not to colorless cells can be considered true positive beads. After sequencing, they can be resynthesized and retested or submitted for other assays such as the functional assay described below.
Lathrop et al. (30) recently reported a new method called the “Bead blot” for identifying ligand-protein and protein-protein interactions using OBOC combinatorial peptide libraries. In this method, a peptide library synthesized on chromatography resin beads is incubated with a starting material that contains a target protein for which a ligand is sought. Then the protein-loaded beads are immobilized in a porous matrix, and the proteins are eluted directionally from the beads and captured on a membrane that is superimposed on the beads. The location of the target protein on the membrane is determined by probing the bead blot with various antibodies, and the beads that originally bound the protein are identified and sequenced. The advantages of the “Bead blot” include the ability to select ligands of unpurified protein, including trace proteins present in complex materials, and ligands of multiple proteins under a variety of conditions in a single experiment.
Aggarwal et al. (31) synthesized a random one-bead one-dimer peptide library on a polyethylene glycol acrylamide (PEGA) resin by modifying the one-bead one-compound method and screened the library with a prostate cancer cell line LNCaP. One peptide, QMARIPKRLARH, was found to bind as a dimer to LNCaP cells that had been spiked into the blood, but it did not bind to normal hematopoietic cells.
In addition to the binding assay, functional assays have been developed for the screening of OBOC libraries to identify specific substrates for protein kinases and proteases. Highly porous PEGA resin is used for the peptide library construction because it allows the enzyme to gain access to the bead interior. To identify peptide substrates for protein kinases, the bead library is incubated with [γ-32P]ATP and the protein kinase. The phosphorylated peptide bead library is washed thoroughly and immobilized on a glass plate with agar. The 32P-labeled beads then are detected by autoradiography, and individual radiolabeled (“active”) beads are isolated for microsequencing.
For protease substrate screening, OBOC peptide libraries with fluorescent dye incorporated at the C-terminus and a quencher at the N-terminus have been used. Peptide-beads susceptible to protease cleavage will fluoresce. These fluorescent-labeled beads were isolated for microsequencing under a fluorescent microscope or isolated by a fluorescent-activated bead sorter (e.g., COPASTMBIOBEAD, Union Biometrica, Inc, Somerville, MA). Information about the substrate sequence, the cleavage point, and the degree of cleavage can be obtained in a single screening. Meldal (32) extended the OBOC concept to generate one-bead two-compound (OBTC) peptide libraries. For example, an OBTC library can be generated on a PEGA resin that contains both a library of inhibitors of proteolytic enzymes and a fluorescence-quenched substrate that competes with the inhibitor for binding to the active site of the enzyme.
Thus far, only a few groups have reported on the release of compounds from OBOC libraries for solution-phase assays. An elegant and powerful approach to screen the OBOC libraries is the in situ solution-phase releasable assay, in which the compound-bead libraries are immobilized in a thin layer of agar (33). Compounds from each bead then are released to the vicinity of each bead for a solution-phase assay in the semi-solid. This method works particularly well for identifying antimicrobial and anticancer agents because a zone of growth inhibition around the positive beads can be detected easily. Jayawickreme et al. (34) reported a “cell-based lawn format” that uses an in situ photocleavage method to release the compound. These in situ releasable solution-phase assays have great potential but will require more development before they can be used reliably for drug screening. For example, special solid supports need to be developed so that all compounds will diffuse freely out of all beads into the surrounding media. We recently have developed novel bilayer shell core beads for such purpose (unpublished work). These beads consist of a polystyrene core in which the coding tags reside and a hydrogel shell on which releasable library compounds are attached.
An alternative approach to using solution-phase assays to screen an OBOC library is to release compounds from an individual or small collection of compound beads in a microtiter plate. The released compounds then are subjected to standard solution-phase assays or are used to print multiple replicates of chemical microarrays. To have enough material from one single bead for biologic assays, one may use macrobeads (250-500 pm diameter) or bead aggregates that are prepared by cross-linking the TentaGel resin beads with glutaraldehyde.
Synthetic Libraries that Require Deconvolution
Many existing solution assays can be applied to the peptide libraries made by this approach. As mentioned above, multistep synthesis and screening that identify only one, not necessarily the best, solution motif are performed.
Self-Assembled PNA-Encoded Chemical Microarrays
In the original report, it was used for the screening of protein binding. Like the OBOC method, a whole-cell binding assay also can be applied to the encoded planar chemical microarrays, although this has not been reported.
Synthetic Libraries that use Affinity Chromatography Selection
For the affinity column selection library, a solution-phase peptide library (a mixture) is loaded onto a column with an immobilized target protein. This method has been applied successfully to retrieve an antibody-specific binding peptide from a mixture of peptides. It also was reported to identify peptide motifs for SH2 domains and kinase domains of protein tyrosine kinases.
Elucidation of the Chemical Structure of Active Peptides
An important goal in screening combinatorial libraries is the identification of novel structures that interact with receptor targets of biologic interest. Depending on the methods used to generate the library, the procedures for the structure determination of active peptides identified from screening vary.
Phage-display peptide library
Standard DNA sequencing is used to elucidate the structure of an active peptide. From the DNA sequence, one then can determine the amino acid sequence of the displayed peptide.
Spatially addressable library
The structure of each peptide in the library is known already; therefore, structure determination of the positive lead is not needed.
OBOC combinatorial library
In peptide libraries composed of α-amino acids (including many unnatural α-amino acids), automatic microsequencing by Edman degradation is the method of choice. However, the Edman sequencing method is expensive, is relatively slow, and requires that the N-termini of the peptide are free; plus, the peptide consists of α-amino acids only. For peptides without a free N-terminus (e.g., cyclic peptides that use the N-terminal amino group for cyclization), branched peptides, and peptides with one or more nonsequenceable building blocks (e.g., β and γ-amino acids), the OBOC library synthesis requires different synthetic and chemical encoding strategies. Based on the chemistry used, decoding can be achieved by either Edman microsequencing, mass spectrometry (MS), or gas chromatography. To eliminate the interference of coding tags with the screening, the coding tags should be in the bead interior (Fig. 2a). We recently have developed two simple, yet highly robust, methods to prepare topologically segregated bilayer beads (Fig. 2b) (35, 36). We recently reported a new “ladder-synthesis” approach that combines both “ladder-synthesis” and bilayer bead concepts to encode OBOC nonsequenceable peptide and peptidomimetic libraries (36). An add-in benefit of the new “ladder-synthesis” is that the library beads generated by this method are amenable to both MS and Edman microsequencing if the compound is a sequenceable peptide (Fig. 2c). More recently, Joo et al. (37) used a similar approach for a high-throughput sequence determination of OBOC cyclic peptide library members using partial Edman degradation/MS.
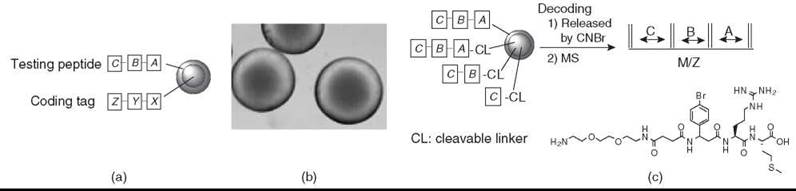
Figure 2. (a) Spatial separation of a testing peptide and coding tag. A, B, and C are sequenceable or nonsequenceable amino acids; X, Y, and Z are sequenceable coding units; and X encodes A, Y encodes B, and Z encodes C. (b) Photomicrograph of topographically segregated bilayer beads. Free amines at the inner core of each bead (which is the site for a coding tag) were reacted with bromophenol blue to show blue color. (c) MS decoding of an OBOC library generated by the new ''ladder-synthesis'' approach.
Synthetic library methods that require deconvolution
Structure determination by physico-chemical methods for deconvolution libraries is not necessary because the chemical structure of the active compound can be deduced from the synthetic history and analysis results of the compound mixtures.
Self-assembled PNA-encoded chemical microarrays
The identity of the positive library compound that interacts with the target protein can be determined by knowing the nucleic acid sequences of the oligonucleotide microarrays.
Affinity column selection
For the synthetic library method that uses the affinity chromatography selection approach, the bound peptides can be eluted and microsequenced by Edman degradation. Concurrent microsequencing of the retrieved peptide mixture can be performed rather than sequencing individual peptides. Sequence motifs then can be defined in a fast and efficient way. However, the amino acid sequence obtained will be the result of the summation of the peptide mixture. Unless a predominant, distinct motif and an alignment of one or more of the critical residues exists within the peptide sequence of the library (e.g., with a fixed residue at a specific position), the result could be very difficult if not impossible to interpret.
Practical Applications of Combinatorial Peptide Libraries
Combinatorial library technology can be employed to study almost any biologic target, and results from these studies can increase our fundamental understanding of cellular function and signaling pathways. It also can be used for the discovery and optimization of drug leads. Random peptide libraries displayed on phages have been used successfully for a variety of biologic applications. These applications include the discovery of peptide ligands to target receptors, specific ligands for DNA sequences, enzyme inhibitors, peptides that mimic carbohydrate structures, protein-protein interfaces, and receptor binding sites. Others include peptide ligands for cancer cells and cancer-associated proteins. In vivo screening of phage-displayed peptide libraries has resulted in the discovery of peptide ligands that bind to the endothelium of tumor blood vessel and lymphatic vessels of specific organs, tissues, or cancers.
Over the last decade, the synthetic combinatorial library approach has been applied successfully to various biologic systems. These systems include the identification of ligands against antibodies (both continuous and discontinuous epitopes), streptavidin, avidin, opioid receptors, melanin-stimulating hormone (MSH) receptors, surface idiotype of B-cell lymphoma cell lines, T cells, bombesin receptors, molecules A2 and B7 of major histocompatibility complex (MHC) class I cytokine receptors, signal transduction adaptor molecules such as SH2 and SH3 domains, adhesion molecules such as gpllb/Illa, specific metal ions, double-stranded DNA, and organic dye molecules. In addition, peptides with antiprotease (including HIV protease), anti-inflammatory, antibacterial, and catalytic activities have been discovered. Furthermore, substrate motifs for various protein kinases, proteases, deacetylases, or other posttranslational modification enzymes have been elucidated. Peptide-based enzyme mimics (artificial enzymes) have been identified through screening combinatorial peptide libraries. We reported the use of a 32P or 33P phosphorylation assay and an autoradiographic method to identify specific and efficient peptide substrates for protein kinases. We also have described the use of whole-cell binding assays in which bead libraries are mixed with live cells to identify cell surface-binding peptide ligands specific for prostate cancer, nonsmall cell lung cancer, and lymphoma cells. Table 2 summarizes some published applications of combinatorial library methods.
Combinatorial chemistry also has been applied successfully to the field of material science (38, 39). This application includes the discovery of polymeric structures with specific physical, chemical, electrochemical, photochemical, or photoelectric properties.
Table 2. Combinatorial library methods and their applications
|
|
Peptide library methods |
Applications (examples) |
|
Biologic library methods |
Phage-display Plasmid-display Polysome-display Fimbria-display Yeast-display Bacterial-display mRNA-display Ribosome-display In vitro selection methods
|
Cell surface ligands (integrins, adhesion molecules or surface receptors, vasculature, tumor lymphatics, normal organs/cells/body fluids); B-cell epitope mapping (gp120, neolactotetraosylceramide); MHC class II molecule binding peptides; peptide substrate for tPA; peptide substrates for HIV-1 protease; and SH2 and SH3domain-binding peptides. |
|
Synthetic library methods |
Spatially addressable parallel library • Multipin technology • SPOT synthesis • Nanokan technology • Peptide microarray in situ synthesis spotting of finished peptides • Multisyringe system • 96-well plate library method
|
B-Cell epitope mapping (foot-and-mouth disease, TMV antigen, shrimp allergen); T-Cell epitope mapping (type 1 diabetes, alloreactive T-cell); protein kinase substrate/inhibitor (protein kinase I); MHC class I molecule binding peptides; and epitope mapping for monoclonal and polyclonal antibodies. |
|
|
Synthetic library methods that require deconvolution • Iterative approach • Positional scanning • Recursive deconvolution approach • Orthogonal partition approach • Dual recursive deconvolution
|
T-Cell epitope mapping (multiple sclerosis, human papillomavirus); B-Cell epitope mapping (various antigens); opiate receptor agonists and antagonists; MHC class I and II molecules binding peptides; protease inhibitors; DNA-binding peptides; mimotopes/epitopes for monoclonal or polyclonal antibodies; Tab-2-binding peptides; antibacterial agents; CAMP-dependent protein kinase motif; and molecules with catalytic and hemolytic activities. |
|
|
One-bead one-compound library method |
B-Cell epitope mapping (insulin); T-Cell epitope mapping (partially cleaved OBOC for diabetes/Tb reactive T-cells); imotopes/epitopes for monoclonal antibodies; Cell surface ligand identification (surface idiotype, integrins); MHC class I binding motif; Protein kinase substrate/inhibitor (cAMP Dependent protein kinase, c-src, c-abl, Etk, metalloproteinase); Protein-binding ligands (SH2 and SH3 domains, avidn, streptavidn, Bombesin-receptor, GpIIb/IIIa, anti-β-endorphin antibody, artificial receptors); Factor Xa inhibitor; Protease substrates and inhibitors; Antibacterial agent; Peptides with catalytic activity
|
|
|
Self-assembled PNA-encoded chemical microarrays |
Protease substrates (serine protease thrombin, cysteine protease caspase-3)
|
|
|
Synthetic library method that requires affinity chromatography selection |
Protein kinase substrate/inhibitor (Csk, 3 bp2, Fps Fes, Grb-2, Hcp, Shc, Syk, Vav, Zap-70); Epitope mapping for monoclonal antibodies; SH2 domain-binding peptides; and kinase domains of protein tyrosine kinases |
Future Directions
Combinatorial chemistry has been playing a central role in the field of chemical biology. Huge libraries of peptides, peptidomimetics, small molecules, natural product-like molecules, and macrocycles can be generated on solid support or in solution phase. Many methods have been developed to screen such libraries. Numerous reports detail the successful application of these technologies to basic research and to the development of pharmaceutics, diagnostics, imaging agents, new materials, catalysts, and biosensors. Biologic libraries such as phage-display peptide libraries, although very powerful and useful, are limited primarily to the standard 20 L-amino acids. With existing technology, the incorporation of a few selected unnatural amino acids into such libraries is, in principle, feasible but far from a routine (40). A need exists to develop highly robust biologic combinatorial chemistry systems that enable one to incorporate many unnatural amino acids, including some that can be modified subsequently through site-specific ligation or derivatization. Peptide and chemical microarrays (planar or bead-based), with or without microfluidics, have been evolving rapidly in the last decade. Many new technologies to generate and screen such microarrays have been developed. One major challenge is to develop a robust label-free optical detector that can measure the real-time binding kinetics of a pure target protein or complex protein mixtures against high-density microarrays (e.g., 10,000-20,000 discrete peptide or chemical spots per three square inches, a size of a microscope glass slide). Another challenge is to develop an efficient in situ light-directed synthesis of chemical molecules other than peptides and nucleic acids in a high-density microarray format.
The OBOC combinatorial library method is highly versatile and economical. It also is a form of chemical microarrays. Many investigators successfully have applied the on-bead screening methods in their research. The solution-phase and cell-based assays for OBOC libraries, however, are much less developed and have been applied successfully in only a few laboratories. A need exists to develop robust methods that allow investigators to screen routinely huge OBOC releasable peptide or chemical libraries (e.g., 200,000 compounds) with multiparametric solution-phase cell-based assays for compounds that affect specific cell signaling pathways or target proteins in one single 10 cm Petri dish.
In the areas of catalysis and material sciences, one would expect that new materials would continue to be developed combinatorially. We will not be surprised if one day novel chlorophyll mimics that consist of peptides and organic chromophores and are developed through combinatory chemistry become a major component of future photocells. The future of combinatorial chemistry is bright, and it will continue to serve as an indispensable research tool for many investigators to solve difficult problems across many different scientific disciplines.
Acknowledgments
The authors would like to thank Dr. Joseph Kappel for editorial assistance. Supported by NIH grants R33 CA89706, NCDDG U19CA113298, R01CA-098116, R01CA15483, and NSF CHE-0302122.
References
1. Geysen HM, Meloen RH, Barteling SJ. Use of peptide synthesis to probe viral antigens for epitopes to a resolution of a single amino acid. Proc. Natl. Acad. Sci. USA 1984; 81:3998-4002.
2. Lam KS, Salmon SE, Hersh EM, Hruby VJ, Kazmierski WM, Knapp RJ. A new type of synthetic peptide library for identifying ligand-binding activity. Nature 1991; 354:82-84.
3. Houghten RA, Pinilla C, Blondelle SE, Appel JR, Dooley CT, Cuervo JH. Generation and use of synthetic peptide combinatorial libraries for basic research and drug discovery. Nature 1991; 354;84-86.
4. Fodor SP, Read JL, Pirrung MC, Stryer L, Lu AT, Solas D. Light-directed spatially addressable parallel chemical synthesis. Science 1991; 25;767-773.
5. Scott JK, Smith GP. Searching for peptide ligands with an epitope library. Science 1990; 249:386-390.
6. Cwirla SE, Peters EA, Barrett RW, Dower WJ. Peptides on phage: a vast library of peptides for identifying ligands. Proc. Natl. Acad. Sci. 1990; 87:6378-6382.
7. Devlin JJ, Panganiban LC, Devlin PE. Random peptide libraries: a source of specific protein binding molecules. Science 1990; 249:404-406.
8. Frank, R. Spot-synthesis: an easy technique for the positionally addressable, parallel chemical synthesis on a membrane support. Tetrahedron 1992; 48:9217-9232.
9. Freier SM, Konings DAM, Wyatt JR, Ecker DJ. Deconvolution of combinatorial libraries for drug discovery: A model system. J. Med. Chem. 1995; 38:344-352.
10. Lam KS, Lebl M, Krchnak V. The “one-bead-one-compound” combinatorial library method. Chem. Rev. 1997; 97:411-448.
11. Winssinger N, Damoiseaux R, Tully DC, Geierstanger BH, Burdick K, Harris JL. PNA-encoded protease substrate microarrays. Chem. Biol. 2004; 11:1351-1360.
12. Songyang Z, Carraway KL, Eck MJ, Harrison SC, Feldman RA, Mohammadi M, Schlessinger, J, Hubbard SR, Smith DP, Eng C, Lorenzo MJ, Ponder BAJ, Mayer BJ, Cantley LC. Catalytic specificity of protein-tyrosine kinases is critical for selective signalling. Nature 1995; 373:536-539.
13. Parmley SF, Smith GP. Antibody-selectable filamentous fd phage vectors: Affinity purification of target genes. Gene (Amsterdam). 1988; 73:305-318.
14. Dani M. Biological libraries. J. Receptor Signal Transduction Res. 2001; 21:447-468.
15. Houghten RA. General method for the rapid solid-phase synthesis of large Numbers of peptides: specificity of antigen-antibody interaction at the level of individual amino acids. Proc. Nat. Acad. Sci. 1985; 82:5131-5135.
16. Nicolaou KC, Xiao XY, Parandoosh Z, Senyei A, Nova MP. Radiofrequency encoded combinatorial chemistry. Angew. Chem. Int. Ed. 1995; 34:2289-2291.
17. Moran EJ, Sarshar S, Cargill JF, Shahbaz, MM, Lio, A, Mjalli, AMM, Armstrong, RW. Radio frequency tag encoded combinatorial library method for the discovery of tripeptide-substituted cinnamic acid inhibitors of the protein tyrosine phosphatase Ptp1b. J. Am. Chem. Soc. 1995; 117:10787-10788.
18. Furka A, Sebestyen F, Asgedom M, Dibo G. General method for rapid synthesis of multicomponent peptide mixtures. Int. J. Pept. Protein Res. 1991; 37:487-493.
19. Gao X, Pellois JP, Na Y, Kim Y, Gulari E, Zhou X. High density peptide microarrays. In situ synthesis and applications. Mol. Divers. 2004; 8:177-187.
20. Gao X, Zhou X, Gulari E. Light directed massively parallel on-chip synthesis of peptide arrays with t-Boc chemistry. Proteomics 2003; 3:2135-2141.
21. Yao W, Hong A, Zhou X, Gao X. Systematic approach to the development of photogenerated bases for parallel synthesis of peptide microarray. In Proc. Abstracts, 62nd Southwest Regional Meeting of the American Chemical Society, Houston, TX, 2006.
22. Xu Q, Miyamoto S, Lam KS. A novel approach to chemical microarray using ketone-modified macromolecular scaffolds: Application in micro cell-adhesion assay. Mol. Divers. 2004; 8:301-310.
23. Dikmans A, Beutling U, Schmeisser E, Thiele S, Frank R. SC2: a novel process for manufacturing multipurpose high-density chemical microarrays. QSAR & Comb. Sci. 2006; 25:1069-1080.
24. Geysen HM, Rodda SJ, Mason TJ. A priori delineation of a peptide which mimics a discontinuous antigenic determinant. Mol. Immunol. 1986; 23:709-716.
25. Pinilla C, Appel JR, Blanc P, Houghten RA. Rapid identification of high affinity peptide ligands using positional scanning synthetic peptide combinatorial libraries. Bio Techniques. 1992; 13:901-902, 904-905.
26. Zhu X Landry JP, Sun YS, Gregg JP, Lam KS, Guo X. Oblique-incidence reflectivity difference microscope for label-free high-throughput detection of biochemical reactions in a microarray format. Appl. Optics. 2007; 46:1890-1895.
27. Grosjean L, Cherif B, Mercey E, Roget A, Levy Y, Marche PN, Villiers MB, Livache T. A polypyrrole protein microarray for antibody-antigen interaction studies using a label - free detection process. Anal. Biochem. 2005; 347:193-200.
28. Krag DN, Shukla GS, Shen GP, Pero S, Ashikaga T, Fuller S, Weaver DL, Burdette-Radoux S, Thomas C. Selection of tumorbinding ligands in cancer patients with phage display libraries. Cancer Res. 2006; 66:7724-7733.
29. Aina OH, Liu R, Sutcliffe JL, Marik J, Pan CX, Lam KS. From combinatorial chemistry to cancer targeting peptides. Mol. Pharmaceutics. 2007; 4:631-651.
30. Lathrop JT, Fijalkowska, I, Hammond D. The Bead blot: A method for identifying ligand-protein and protein-protein interactions using combinatorial libraries of peptide ligands. Anal. Biochem. 2007; 361:65-76.
31. Aggarwal S, Harden J, Denmeade Sr. Synthesis and screening of a random dimeric peptide library using the one-bead-one-dimer combinatorial approach. Bioconjugate Chem. 2006; 17:335-340.
32. Meldal M. The one-bead two-compound assay for solid phase screening of combinatorial libraries. Biopolymers 2002; 66:93-100.
33. Salmon SE, Liu-Stevens RH, Zhao Y, Lebl M, Krchnak V, Wertman K, Sepetov N, Lam KS. High-volume cellular screening for anticancer agents with combinatorial chemical libraries: a new methodology. Mol. Divers. 1996; 2:57-63.
34. Jayawickreme CK, Sauls H, Bolio N, Ruan J, Moyer M, Burkhart W, Marron B, Rimele T, Shaffer J. Use of a cell-based, lawn format assay to rapidly screen a 442,368 bead-based peptide library. J. Pharmacol. Toxicol. Methods 1999; 42:189-197.
35. Liu R, Marik J, Lam KS. A novel peptide-based encoding system for “one-bead one-compound” peptidomimetic and small molecule combinatorial libraries. J. Am. Chem. Soc. 2002; 124:7678-7680.
36. Wang X, Peng L, Liu R, Gill SS, Lam KS. Partial allocdeprotection approach for ladder synthesis of “one-bead one-compound” combinatorial libraries. J. Comb. Chem. 2005; 7:197-209.
37. Joo SH, Xiao Q, Ling Y, Gopishetty B, Pei D. High-throughput sequence determination of cyclic peptide library members by partial edman degradation/mass spectrometry. J. Am. Chem. Soc. 2006; 128:13000-13009.
38. Berkessel A. The discovery of catalytically active peptides through combinatorial chemistry. Curr. Opi. Chem. Biol. 2003; 7:409-419.
39. Hoveyda AH. Catalyst discovery through combinatorial chemistry. Chem. biol. 1998; 5:187-191.
40. Tsao ML, Tian F, Schultz P. Incorporation of unnatural reactive amino acids into phage display libraries using orthogonal mutant aminoacyl-tRNA synthetases for post-translational modification. PCT Int. Appl. 2007; 114.
Further Reading
Krumpe LRH, Mori T. Potential of phage-displayed peptide library technology to identify functional targeting peptides. Expert Opi. Drug Discovery 2007; 2:525-537.
Liu R, Wang X, Song A, Bao T, Lam KS. Development and applications of topologically segregated bilayer beads in one-bead one-compound combinatorial libraries. QSAR & Comb. Sci. 2005; 24:1127-1140.
Marik J, Lam KS. Peptide and small-molecule microarrays. Methods Mol. Biol. 2005; 310:217-226.
Marik J, Xu Q, Wang X, Peng L, Lam KS. A novel encoded high-density chemical microarray platform for proteomics and drug development. pp. 849-853 In: Peptides-Peptide Revolution: Genomics, Proteomics & Therapeutics, Proc. 18th American Peptide Symposium. Chorev, Michael and Tomi K. Sawyer, eds. 2004. American Peptide Society, San Diego, California.
Prepare peptide microarray using digital chemistry: http://gaolab.chem.uh.edu/res1.htm
Samoylova TI, Morrison NE, Globa LP, Cox NR. Peptide phage display: opportunities for development of personalized anti-cancer strategies. Anti-Cancer Agents Med. Chem. 2006; 6:9-17.
Wenschuh H, Gausepohl H, Germeroth L, Ulbricht M, Matuschewski H, Kramer A, Volkmer-Engert R, Heine N, Ast T, Scharn D, Schneider-Mergener, J. Positionally addressable parallel synthesis on continuous membranes. In: Combinatorial Chemistry. Fenniri, Hicham, ed. 2000. Oxford University Press, Oxford, UK.
See Also
Chemical Ligation: Peptide Synthesis
Peptide Synthesis
Peptides, Chemistry of
Phage Display
Synthetic Peptides and Proteins to Elucidate Biological Function