CHEMICAL BIOLOGY
Locked Nucleic Acids
Zenia M. Sterling and Troels Koch, Santaris Pharma A/S, Harshoim, Denmark
doi: 10.1002/9780470048672.wecb516
The intensive research and development of antisense oligonucleotides in the past decades has refined our knowledge of this class of molecules, and they are now recognized as potent reagents for gene-target validation and therapeutic uses. Within this field, a very promising class is locked nucleic acid (LNA). In recent years, the number of applications for LNA in basic research, molecular diagnostics, and therapeutics has expanded continuously. In this article, the general biophysical, biochemical, and pharmacological properties of LNA will be summarised. The ability of LNA oligonucleotides to target both coding and noncoding RNAs effectively and specifically via different modes of action will be discussed. Several LNA oligonucleotides have completed preclinical toxicology and are now tested in clinical trials. The status of one drug candidate will be presented briefly at the end of the article.
The principle of antisense was first described in 1978 by Zamecnik and Stephenson (1). In antisense gene expression is inhibited at the translational level by sequence specific hybridization of an oligonucleotide to mRNA, and thereby inhibiting the expression of the protein encoded gene. Theoretically, the antisense principle can be used to treat any disease caused by the expression of a deleterious gene, and as hypothesized by Zamecnik and Stephenson, this principle is very likely to have big potential in therapeutics. However, it turned out to be harder to produce effective antisense drugs than anticipated, but recent discoveries and developments in the area have changed the picture, and the prospects of antisense seems brighter than ever.
Background
Native oligonucleotides can not be used as active antisense reagents and therefore in the history of antisense, a variety of chemically modified nucleotides has been developed to improve properties such as target affinity, nuclease resistance, and pharmacokinetics of the oligonucleotides. The first generation of chemically modified antisense drugs was the phosphorothioates (PSs). The phosphorothioate internucleoside link showed improved nuclease stability and bioavailability, but it resulted in lower RNA affinity. To compensate for that, the length of PS was kept rather long and 18-20-mers were frequently used. This is significantly longer than the antisense principle requires for specific mRNA recognition. The ideal antisense reagent ought not to be longer than required for specific mRNA recognition which is-dependent on sequence-typically 12-16-mers. However, PSs have been used extensively to inhibit mRNA targets in vitro, but the combination of a relatively low target affinity and limited nuclease stability reduce the in vivo potential of PS via specific antisense mechanisms. On the contrary, the therapeutic significance of PS is high in combination with other chemical modifications (see below) or via their mode of action other than classical antisense.
The second-generation antisense drugs are represented classically by the nucleotides where the 2'position has been modified. Examples of this class are the 2’-O-Me, 2’-O-MOE, and 2’-F-modified nucleotides (Fig. 1)
By incorporating these modifications into oligonucleotides, the affinity was improved—compared with the PSs—but to get sufficient potency and efficacy, the length of the second generation antisense drugs was unchanged compared with the first generation (18-20-mer) drugs illustrating that the second generation antisense oligonucleotide did not increase the affinity adequately.
Locked Nucleic Acid (LNA) was presented in 1998. LNA represents a true member of the third generation of antisense drugs. When LNA nucleotides were incorporated into oligonucleotides, they induced the highest binding affinity for both complementary RNA and DNA reported in the field (2). This opened completely new perspectives for the development of antisense drugs. The remaining part of this article will focus on LNA and its therapeutic aspects.
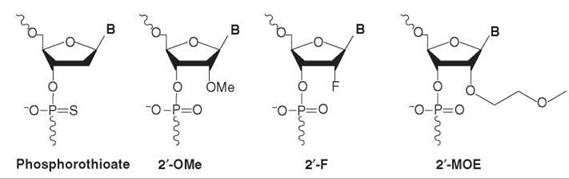
Figure 1. Molecular structure of examples of first- and second-generation antisense drugs.
Basic Properties of LNA
LNA is defined as an oligonucleotide comprising one or more 2'-O,4'-C-methylene-β-D-ribofuranosyl nucleotide building blocks (2). The term LNA was selected to signal the structural fact that the bicyclic structure of LNA nucleoside locks conformational flexibility of the ribose ring. Furthermore, it turned out that bicyclic structure of the LNA-monomer served as a general template for development of additional analogs of LNA with additional useful properties. Examples of such LNA analogs are amino-/thio- and α-L-LNA where the 2'-,4'-bicyclic structure also is composed of five-membered rings (Fig. 2)
The design rationale for making the 2'-O,4'-C-oxymethylene link was to make a high-affinity RNA analog by preorganizing—locking—furanose in a North-type conformation. The molecular consequences of doing this was thought to be twofold: 1) It would ensure that nucleotides were preorganized in a high-affinity form (North), and 2) the rigid structure of the bicyclic nucleotides would make hybridization less entropically unfavorable (2, 3). LNA nucleotides are always hybridizing in a locked North-like structure, which is an important contribution to the affinity gain, but the positive entropy contribution is not the entire explanation, from a thermodynamically point of view the full picture of affinity increase was more complex (4, 5, 6). It has now been realized that there are many contributing components of which stacking, rational freedom of nucleotides/oligonucleotides, structure of helices, water re- lease/binding, and so on are important elements.

Figure 2. Molecular structure of LNA and LNA analogs.
Biophysical Properties of LNA
LNA oligonucleotides obey Watson-Crick hydrogen binding rules and form right-handed helices, and actually the most intriguing property of the LNA nucleotide is that it fits perfectly the Watson-Crick framework. LNA nucleotides can be included in any combination with DNA/RNA nucleotides, and any analog hereof that fits within the Watson-Crick framework.
The immediate property described for LNA was an extraordinary high affinity (2)—a property later confirmed in numerous papers and recently reviewed (7). When LNA monomers are incorporated in either DNA or RNA each residue adds to affinity in an additive manner. Insertions of few LNA residues lead to increase of melting temperature (Tm) of 6-7 ° C per modification; however, the largest increase is observed when LNA nucleotides are included in PS, which provides a record high increase of 9-10 °C per modification (8). This feature of LNA is unique because PS modification is known to reduce the affinity of DNA and 2'-modified oligonucleotide analogs. However, when PSs are used as internucleoside link in LNA oligonucleotides affinity is almost not affected and therefore, the affinity of LNA diesters and LNA PSs is almost the same.
The affinity increase per LNA modification is the greatest when only few LNA nucleotides are incorporated into oligonucleotides because LNA nucleotides direct the conformation of nearby deoxyribose-nucleotides to attain a greater degree of the high affinity North conformation. Therefore, the affinity increase is driven by both the LNA nucleotides and the North-directed DNA nucleotides. Fully modified LNA oligonucleotides—with no DNA nucleotides—do have the highest affinity, but the affinity per LNA modification is lower. This unique property of LNA is called “structural saturation.” Structural saturation is a very important property of LNA because only a few LNA modifications can transform the entire oligonucleotide into a high-affinity structure.
The high affinity of LNA makes it possible to design shorter antisense oligonucleotides with retained high potency, and it means that the “hit rate” of LNA antisense oligonucleotides designed to complement different sequences along the mRNA are greater compared with traditional antisense chemistries (9). This finding indicates that the lead identification process in drug discovery becomes faster and that fewer LNA antisense oligonucleotides are needed to find highly potent mRNA binders.
LNA and most LNA-analogs can be mixed in any combination with nucleic acids and analogs. Depending on the application of LNA, the design may be divided in five categories: mixmer, gapmer, headmer, tailmer, and fully modified (Fig. 3, a-e) In the mixmer, the LNA residues are dispersed along the sequence of the oligonucleotide (a), whereas in the gapmer two continuous LNA segments are attached in the 5'- and 3'-ends of a central nucleic acid fragment (b). A headmer is a continuous nucleic acid fragment with a continuous LNA segment in the 5'-end (c) and vice versa for a tailmer (d). The fully modified LNA oligonucleotide is self-explanatory (e) (Fig. 3).
The length and specific design, however, may vary depending on the intended use and on how important the affinity, nuclease resistance, and the RNase H/RISC recruitment (see below) is for the proposed application. In the classic designs described above, the total length of LNA oligonucleotides is 16 mers, and the oligonucleotides are fully PS-modified.
In conclusion, freedom of design offered by LNA has enabled the development of LNA oligonucleotides with the ability to target both coding and noncoding RNAs effectively and specifically in vivo using different modes of action, such as target cleavage, steric block of translation and inactivation by sequestration.

Figure 3. General designs of LNA antisense oligonucleotides.
One important aspect of antisense technology is the stability of antisense agents against extracellular and intracellular endonucleases and exonucleases. A series of studies has investigated the serum and nuclease stability of LNA-modified oligonucleotides, and the studies have shown that incorporation of LNA segments into oligonucleotides increase their stability against nucleases. It is the number and the position of the LNA residues as well as insertion of other stabilizing entities—such as PSs—that determines the rate of degradation of the oligonucleotides. For example, a single LNA residue at the 3'-end is not sufficient to increase stability substantially against exonucleolytic attack, whereas one LNA residue at the 3'-penultimate position or two LNA modifications at the ultimate position of the 3'-end efficiently stabilize against 3'-endonucleases (10, 11). An effective way to secure exonucleolytic protection is obtained with the gapmer design, but the central DNA segment is sensitive to endonucleolytic activity. If the central DNA segment becomes longer than four nucleotides, then cleavage starts to occur and the cleavage rate increases as the DNA segment increases.
A preferred way to protect degradation of the central DNA segments is to use PS internucleoside links. The PS link is easy to introduce during LNA synthesis and it serves as a substrate for RNase H (see below). A series of fully PS-modified gapmers with gap-sizes that range from 9 to 11 PS links have been examined in human and rat plasma and they showed no sign of endonucleolytic activity (7). Fully modified LNA is essentially resistant to exonucleolytic and endonucleolytic degradation either with fully diester or PS internucleoside links (11).
RNase H recruitment
It is well established that oligonucleotides targeting mRNA are most potent if they are designed to recruit RNase H. Under normal circumstances, this enzyme cleaves mRNA hybridized to a DNA complement. Nevertheless, because LNA nucleotides adopt an RNA-like structure, they cannot recruit RNase H. However, LNA gapmers, which contain a stretch of DNA—or PS—nucleotides, can recruit RNase H. More specifically, several studies has investigated various LNA/DNA gapmers and concluded that a gap of six DNA nucleotides is necessary for initiating RNase H activity, and that a gap of seven DNA nucleotides allows for high RNase H activity (11-14). The conclusion is that antisense LNA oligonucleotides can be designed to elicit RNase H activity while still containing LNA monomers for improved binding and target accessibility (15). Furthermore, it has been shown that the highest activity was observed if the entire LNA gapmer was phosphorothioate (not just the DNA segment) (11)
LNA Applied In Vitro
Most antisense experiments made with LNA have been focused mechanistically on mRNA inhibition by RNase H recruitment (7). Expression studies in a wide variety of human epithelial and cancer cell lines are made and the overall picture of these experiments is that LNA antisense oligonucleotides are very potent with IC50 values for mRNA inhibition obtained frequently in the low to sub nanomolar range—and actually more potent than competing oligonucleotide analogs (7).
The potency of LNA oligonucleotides is related to the design of the oligonucleotide and to the target site. Several studies have found that the gapmer design is slightly more potent than the mixmer design, in which inhibition of mRNA in the translational start site is required (7). However, when the target site is in the coding region, the gapmer design is superior (16). This finding is caused by the fact that a high-affinity LNA mixmer can prevent ribosome binding to mRNA and thereby stop translation initiation; but once the ribosome is bound and initiates translation, mRNA has to be cleaved by RNase H to stop its action (17). LNA gapmers have been used intensively in cellular-based assays to inhibit mRNA expression. This work has been reviewed recently (7), and the conclusion is that LNA gapmers ranging from 12-16-mers inhibits gene expression with IC50 values in the low to sub-nanomolar range (18).
Another mechanistic action for mRNA inhibition is a nonR-Nase H based mechanism in which the LNA antisense oligonucleotide acts via very tight binding to the mRNA and thus, interferes with its additional processing. This mechanism is called “steric block.” Steric block includes prevention of ribosome binding by blocking the 5'-UTR that acts as a “road block” during translation by binding to the message or by redirection of splicing sites to alter the production of splice variants (19). When antisense oligonucleotides act according to this mechanism, the mRNA remains intact. LNA mixmers have been used effectively as steric blocks to inhibit the expression of several therapeutically interesting targets for instance inhibition of viral replication (11, 20) and redirect splicing in samples from patients with Duchenne muscular dystrophy (21, 22).
Pharmacology
Currently, pharmacological activity of LNA antisense oligonucleotides has been demonstrated in many different tissues (liver, kidney, colon, jejunum, small intestine, lung, spleen, and brain) and species (mice, rats, and monkeys) using a variety of administration routes (subcutaneous infusion, intravenous injection, intraperitoneal injection, and direct injection into brain) (7). Using different designs of LNA oligonucleotides, it has been possible to target both mRNAs and noncoding miRNAs effectively (see below), which causes either a reduction in protein synthesis, a shift in the splicing pattern, or inactivation by biophysical sequestration.
The pharmacological effect of LNA oligonucleotides depends on biodistribution and cellular uptake of the LNA and LNA-analog oligonucleotides. Biodistribution studies have shown equal uptake of LNA and α-L-LNA, except in kidneys, where the uptake of α-L-LNA is greater than for LNA. Amino- and thio-LNA have increased uptake in liver, whereas the amino-LNA has the broadest distribution showing significantly greater uptake in heart, lung, muscle, and bone compared with other analogs (23). Once absorbed by tissues, LNA oligonucleotides are taken up by cells by internalization, and a strong relationship between tissues up-take and endogenous effects have been demonstrated (7).
The first in vivo antisense experiment with LNA-antisense involved two different 15-mer LNA designs that target the delta opioid receptor (DOR) mRNA in the central nervous system of living rats (14). On direct injection of the LNA oligonucleotides into the brain, a dose-dependent and highly efficacious knockdown of DOR was induced both for an LNA/DNA mixmer and an LNA/DNA/LNA gapmer. In a later study, a fully modified 16-mer LNA, which targeted the RNA polymerase II gene product, inhibited tumor growth in mice at low doses (less than 5 mg/kg/day) and was tolerated well (24). Since these early studies, the pharmacological properties of LNA have been documented in numerous publications. Many improvements have been made, and the LNA oligonucleotide design and how LNA oligonucleotides interact with the intracellular molecular machinery are better understood. The conclusion of these works is that pharmacological relevant doses of LNA range typically from 0.5-5 mg/kg, and that the oligonucleotides are generally well tolerated (7).
An interesting observation is that the pharmacological potential can be increased with reduced size of the LNA oligonucleotides. A shortmer design (oligonucleotides shorter than 16-mers) has been used to inhibit ApoB-100 expression in liver and jujenum. ApoB-100 is the major cholesterol-carrying protein in low-density lipoprotein (LDL) and plays a central role in cholesterol metabolism and artherosclerosis. The study showed that 12-14-mers reduced the target expression with 80-90% where a reduction of only 30-40% was seen with the 15 and 16-mers. The reduction in ApoB-100 expression was followed almost linearly with reduction in plasma cholesterol levels (25). These data suggest that use of shortmers may become a new approach for systemic gene targeting with significantly improved pharmacological effects.
Inhibition of MicroRNA
MicroRNAs (miRNAs) are an abundant class of short endogenous noncoding RNAs that act as posttranscriptional modulators of gene expression. To date, more than 5000 miRNAs have been identified in invertebrates, vertebrates, and plants according to the miRBase miRNA database release 10.0 in August 2007. Growing evidence shows that miRNAs exhibit a wide variety of regulatory functions and exert significant effects on cell growth, development, and differentiation.
A major obstacle to specific and sensitive detection of mature miRNAs in animals is their small size (~19-23 nucleotides). Endlabeled LNA-probes, in which every third position is an LNA residue, have been used in Northern blot analysis to detect miRNAs with high specificity and showing at least 10-fold increased sensitivity compared with DNA probes (26), which is a major achievement in miRNA research. Furthermore, LNA probes with uniform Tm values of 72 °C have been used in high-sensitivity arrays for miRNA expression profiling (27). Such arrays allow discrimination between single nucleotide variations as demonstrated by the specific discovery of related miRNA family members. LNA probes have been used successfully to enhance the sensitivity and specificity of in situ hybridizations to miRNAs in zebrafish (28, 29) and in human brain (30).
The expanding list of human miRNAs along with their highly diverse expression patterns and high number of potential target mRNAs suggest that miRNAs are involved in a wide variety of human diseases. Perturbed miRNA expression patterns are reported in many human cancers (31), and more than 50% of the human miRNA genes are located in cancer-associated genomic regions or at fragile sites (32).
Several studies have shown that LNA oligonucleotides can be used to inhibit miRNA function specifically and effectively (LNA-antimiRs). One study showed inhibition of miR-21 activity with an LNA gapmer that leads to induction of apoptosis in glioblastoma cell lines, which illustrates that aberrantly expressed miRNAs can function as oncogenes (33). Another study showed inhibition of the bantam miRNA in Drosophila by a complementary LNA-modified oligonucleotide resulting in sequence specific derepression of Hid protein synthesis in cell culture (34). Recent findings by Elmen et al. (35) show that high-affinity antagonism of the liver-specific microRNA 122 (miR-122) in mice, by steric hindrance using LNA-antimiRs, can result in potent functional inhibition. This finding suggests that oligonucleotides composed of LNA may be valuable tools for identifying miRNA targets in vivo and for studying the biological role of miRNAs and miRNA-associated gene-regulatory networks in a physiological context. In addition, the high metabolic stability of LNA-antimiRs, caused in part by increased nuclease resistance, their small size and apparent lack of acute toxicity or changes in liver morphology (35) imply that LNA-antimiRs may be well suited as a novel class of potential therapeutics for disease-associated microRNAs.
LNA Drugs in Clinical and Preclinical Development
In 2006, the LNA oligonucleotide SPC2996 completed its first human clinical trial, which was a Phase I/II study in patients with chronic lymphocytic leukemia (CLL). SPC2996 acts by inhibiting the synthesis of Bcl-2, which is a key sensor protein that protects cells against apoptosis. The protein is expressed in most cancers including CLL, and high expression levels have been firmly correlated with low response rates, resistance to chemotherapy, faster time to relapse, and shorter survival time (36). Thus, downregulation of Bcl-2 expression provides an attractive means by which CLL and other cancers can be resensitized to natural apoptotic stimuli and to chemotherapeutic agents that provoke apoptosis.
The results of this study are encouraging for such an early stage trial. All patients who received the top dose of the drug had a rapid reduction in the number of cancerous white blood cells in circulation, and expression of Bcl-2 was reduced with 33% as measured by QPCR in total blood RNA.
The results of this first human clinical trial provide the basis for additional development of SPC2996, and it underlines that repeated doses of the drug can be administered safely.
The second LNA oligonucleotide that entered a human clinical Phase I/II trial in patients with solid tumors is ENZ2968—a potent inhibitor of Hif-1α. Hif-1α serves as the key sensor of cellular hypoxia in response to which it transcriptionally upregulates a host of genes that play important roles in promotion of cancers, which include angiogenesis, apoptosis, cell migration, and metastasis. Hif-1a is essentially undetectable in normal cells, but it reaches high intracellular concentrations in a variety of cancers in which it is correlated strongly with poor prognosis and resistance to therapy.
SPC3833 is in preclinical development and it is designed to effectively alter the expression of apolipoprotein b100 (ApoB100). The drug effectively down-regulate ApoB-100 synthesis leading to a reduction of VLDL secretion from the liver. The down-stream effect of this is that SPC3833 efficiently lowers LDL and cholesterol levels (37). Finally, SPC3649 is the first LNA oligonucleotide developed as a microRNA inhibitor. SPC3649 is a lead candidate targeted to the liver-specific microRNA-122. The LNA antimiR is developed for treatment of Hepatitis C and hypercholesterolemia and is expected to commence human clinical pharmacology and safety studies in 2008.
Final Remarks
The remarkable hybridization properties of LNA, both with respect to affinity and specificity, position LNA as an enabling molecule for molecular biology research, diagnostic applications, and therapeutics. The fact that LNA nucleotides can be mixed in any combination with DNA/RNA residues and phosphorothioate internucleoside linkages is a very attractive property. Of particular relevance for the therapeutic aspects of LNA is the combination of LNA/DNA residues and PS’s. These “chimeric” oligonucleotides exert their action via the individual components: The LNA residue mediates very high target affinity and biostability, and the PS’s provide the necessary PK properties and cellular uptake. We believe that the combination of these properties in one oligonucleotide—as we have described here—provides enabling properties of LNA as RNA inhibitor.
References
1. Zamecnik PC, Stephenson ML. Inhibition of Rous sarcoma virus replication and cell transformation by a specific oligodeoxynucleotide. Proc. Natl. Acad. Sci. U.S.A. 1978; 75:280-284.
2. Singh SK, Nielsen P, Koshkin A, Wengel J. LNA (locked nucleic acids): Synthesis and high-affinity nucleic acid recognition. Chem. Commun. 1998; 455-456.
3. Singh SK, Wengel J. Universality of LNA-mediated high-affinity nucleic acid recognition. Chem. Commun. 1998; 1247-1248.
4. Jensen GA, Singh SK, Kumar R, Wengel J, Jacobsen JP. A comparison of the solution structures of an LNA:DNA duplex and the unmodified DNA:DNA duplex. J. Chem. Soc. Perkin. Trans. 2001; 2:1224-1232.
5. Obika S, Nanbu D, Hari Y, Morio JAK, Doi T, Imanishi T. Stability and structural features of the deplexes containing nucleoside analogues with a fixed N-type conformation, 2'-O,4'-C-methyleneribonucleosides. Tetrahedron Lett. 1998; 39:5401-5404.
6. Koshkin A, Nielsen P, Meldgaard M, Rajwanshi VK, Singh SK, Wengel J. LNA (locked nucleic acid): an RNA mimic forming exceedingly stable LNA: LNA Duplexes. J. Am. Chem. Soc. 1998; 120:13252-13253.
7. Koch T, Oerum, H. Locked nucleic acids. In: Antisense Drug Technologies: Principals, Strategies, and Applications. 2nd edition. Crooke ST, ed. 2007. Taylor & Francis Group, London, UK. pp. 519-564.
8. Bondensgaard K, Petersen M, Singh SK, Rajwanshi VK, Kumar R, Wengel J, Jacobsen JP. Structural studies of LNA: RNA duplexes by NMR: conformations and implications for RNase H activity. Chem. Eur. J. 2000; 6:2687-2695.
9. Grunweller A, Wyszko E, Bieber B, Jahnel R, Erdmann VA, Kurreck J. Comparison of different antisense strategies in mammalian cells using locked nucleic acids, 2'-O-methyl RNA, phos- phorothioates and small interfering RNA. Nucleic Acids Res. 2003; 31:3185-3193.
10. Di Giusto D, King GC. Strong positional preference in the interaction of LNA oligonucleotides with DNA polymerase and proofreading exonuclease activities: implications for genotyping assays. Nucleic Acids Res. 2004; 32(3) 32.
11. Frieden M, Christensen SM, Mikkelsen ND, Rosenbohm C, Thrue CA, Westergaard M, Hansen HF, Orum H, Koch T. Expanding the design horizon of antisense oligonucleotides with alpha-L-LNA. Nucleic Acids Res. 2003; 31:6365-6372.
12. Elmen J, Zhang HY, Zuber B, Ljungberg K, Wahren B, Wahlestedt C, Liang Z. Locked nucleic acid containing antisense oligonucleotides enhance inhibition of HIV-1 genome dimerization and inhibit virus replication. FEBS Lett. 2004; 578:285-290.
13. Kurreck J, Wyszko E, Gillen C, Erdmann VA. Design of antisense oligonucleotides stabilized by locked nucleic acids. Nucleic Acids Res. 2002; 30:1911-1918.
14. Wahlestedt C, Salmi P, Good L, Kela J, Johnsson T, Hokfelt T, Broberger C, Porreca F, Lai J, Ren K, Ossipov M, Koshkin A, Jakobsen N, Skouv J, Oerum H, Jaconsen MH, Wengel J. Potent and nontoxic antisense oligonucleotides containing locked nucleic acids. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:5633-5638.
15. Jepsen JS, Wengel J. LNA-antisense rivals siRNA for gene silencing. Curr Opin. Drug Discov. Devel. 2004; 7:188-194.
16. Braasch DA, Liu Y, Corey DR. Antisense inhibition of gene expression in cells by oligonucleotides incorporating locked nucleic acids: effect of mRNA target sequence and chimera design. Nucleic Acids Res. 2002; 30:5160-5167.
17. Jepsen JS, Pfundheller HM, Lykkesfeldt AE. Downregulation of p21(WAF1/CIP1) and estrogen receptor alpha in MCF-7 cells by antisense oligonucleotides containing locked nucleic acid (LNA). Oligonucleotides 2004; 14:147-156.
18. Arzumanov A, Stetsenko DA, Malakhov AD, Reichelt S, Sorensen MD, Babu BR, Wengel J, Gait MJ. A structure-activity study of the inhibition of HIV-1 Tat-dependent trans-activation by mixmer 2'-O-methyl oligoribonucleotides containing locked nucleic acid (LNA), alpha-L-LNA, or 2'-thio-LNA residues. Oligonucleotides 2003; 13:435-453.
19. Kole R, Weissman SM. Accurate in vitro splicing of human beta-globin RNA. Nucleic Acids Res. 1982; 10:5429-5445.
20. Nulf CJ, Corey D. Intracellular inhibition of hepatitis C virus (HCV) internal ribosomal entry site (IRES)-dependent translation by peptide nucleic acids (PNAs) and locked nucleic acids (LNAs)1.Nucleic Acids Res. 2004; 32:3792-3798.
21. Aartsma-Rus A, Kaman WE, Bremmer-Bout M, Janson AA, den Dunnen JT, van Ommen GJ, van Deutekom JC. Comparative analysis of antisense oligonucleotide analogs for targeted DMD exon 46 skipping in muscle cells. Gene Ther. 2004; 11:1391-1398.
22. Elayadi AN, Braasch DA, Corey DR. Implications of high-affinity hybridization by locked nucleic acid oligomers for inhibition of human telomerase. Biochemistry 2002; 41:9973-9981.
23. Fluiter K, Frieden M, Vreijling J, Rosenbohm C, De Wissel MB, Christensen SM, Koch T, Orum H, Baas F. On the in vitro and in vivo properties of four locked nucleic acid nucleotides incorporated into an Anti-H-Ras antisense oligonucleotide 1. Chem-BioChem. 2005; 6:1104-1109.
24. Fluiter K, Ten Asbroek AL, De Wissel MB, Jakobs ME, Wissenbach M, Olsson H, Olsen O, Oerum H, Baas F. In vivo tumor growth inhibition and biodistribution studies of locked nucleic acid (LNA) antisense oligonucleotides. Nucleic Acids Res. 2003; 31:953-962.
25. Rosenbohm C, Hansen JB, Fisker N, Hedtjarn M, Ravn J, Meldgaard M, Hansen HF, Straarup EM. The effect of shortening locked nucleic acid antisense oligonucleotides, from 16 to 12 nucleotides, on the expression of apoB mRNA and serum cholesterol levels (abstract). J. Clin. Lipidol. 2007; 1:365.
26. Valoczi A, Hornyik C, Varga N, Burgyan J, Kauppinen S, Havelda Z. Sensitive and specific detection of microRNAs by northern blot analysis using LNA-modified oligonucleotide probes. Nucleic Acids Res. 2004; 32:175.
27. Castoldi M, Schmidt S, Benes V, Noerholm M, Kulozik AE, Hentze MW, Muckenthaler MU. A sensitive array for microRNA expression profiling (miChip) based on locked nucleic acids (LNA). RNA 2006; 12:913-920.
28. Kloosterman WP, Wienholds E, de Bruijn E, Kauppinen S, Plasterk RH. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat. Methods 2006; 3:27-29.
29. Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science 2005; 309:310-311.
30. Nelson PT, Baldwin DA, Kloosterman WP, Kauppinen S, Plasterk RH, Mourelatos Z. RAKE and LNA-ISH reveal microRNA expression and localization in archival human brain. RNA 2006; 12:187-191.
31. Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Alder H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:15524-15529.
32. Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:2999-3004.
33. Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005; 65:6029-6033.
34. Orom UA, Kauppinen S, Lund AH. LNA-modified oligonucleotides mediate specific inhibition of microRNA function. Gene 2006; 372:137-141.
35. Elmen J, Lindow M, Silahtaroglu, Bak M, Christensen M, Lind-Thomsen A, Hedtjarn M, Hansen JB, Hanse HF, Straarup EM, McCullagh K, Kearney P, Kauppinen S. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2008; 36:1153-1162.
36. Shangary S, Johnson DE. Recent advances in the development of anticancer agents targeting cell death inhibitors in the Bcl-2 family. Leukemia 2003; 17:1470-1481.
37. Bell TA, III Brown JM, Graham MJ, Lemonidis KM, Crooke RM, Rudel LL. Liver-specific inhibition of acyl-coenzyme a:cholesterol acyltransferase 2 with antisense oligonucleotides limits atherosclerosis development in apolipoprotein B100-only low-density lipoprotein receptor-/-mice. Arterioscler Thromb. Vasc. Biol. 2006; 26:1814-1820.
MicroRNA (miRNA)
mRNA Localization and mRNA Levels, Control of
Oligonucleotide-Directed Inhibition of Gene Expression
Drug Discovery
DNA-Based Structures, Applications of Chemical Biology