CHEMICAL BIOLOGY
Noncoding RNAs
Allison A. Henry and JOrg Vogel, Max Planck Institute for Infection Biology, Berlin, Germany
doi: 10.1002/9780470048672.wecb522
After decades of being either underestimated as a simple messenger in the expression of proteins from deoxyribonucleic acid (DNA), or revered as the molecule from which all life potentially originated, ribonucleic acid (RNA) is at last enjoying a well-deserved golden age as modern techniques allow researchers to uncover the myriad of subtle roles that this molecule plays in the cells of today. Noncoding RNAs, by definition, do not become translated into protein. This broad category includes cellular RNAs that range in size from 20 to 20,000 nucleotides, can contain chemical modifications, adopt a wide variety of secondary and tertiary structures, associate with proteins and other RNA molecules, and have physiological roles, in all organisms, that run the gamut from catalysis to regulating gene expression. As this article is intended to complement the other RNA-related sections of this volume, a brief introduction to all types of noncoding RNA will be followed by a comprehensive discussion of three important categories: riboswitches, small RNAs in bacteria, and large RNAs in mammals. Riboswitches are cis-acting RNA elements that modulate gene expression in a highly specialized manner by means of an aptamer domain, an RNA sequence that tightly and selectively binds a given metabolite. Small RNAs are 70-200 nucleotide molecules in bacteria that regulate gene expression, primarily at a posttranscriptional level. In mammalian cells, large RNAs of up to 19,000 nucleotides are essential to X-chromosome inactivation and genomic imprinting.
To a chemist, who is accustomed to thinking about the enormous diversity of structures and functions to be found in small molecules, ribonucleic acid (RNA) may seem humdrum. It is, after all, a polymer composed of only four subunits, which comprise a ribose ring and a purine or pyrimidine, that are linked together by phosphodiesters. From a simpleminded perspective, RNA has fivefold fewer types of functionality than proteins, which are made up of 20 different residues; in fact, RNA is similar in composition to deoxyribonucleic acid (DNA), the preferred molecule of genetic information storage that does not have a reputation for carrying out interesting chemistry inside the cell. Such an observer would likely be content with the notion that the roles of RNA are limited to coding for proteins and serving as viral genomes, in accordance with the biological dogma that existed until about three decades ago. Since then, new technologies have enabled significant progress in our understanding of the diversity of structures and functions of RNA molecules that exist in the cells of all organisms. Concomitant with our discovery and characterization of new RNA molecules, the nomenclature in the field has expanded, and for the sake of clarity, we risk stating the obvious: RNA that codes for protein is referred to as messenger RNA (mRNA) and all others are noncoding RNAs, by definition. Three categories of noncoding RNA, riboswitches, regulatory small RNAs in bacteria (sRNAs), and large noncoding RNAs in mammals, will be discussed in some detail in this article, but first a summary of the features and functional repertoire of noncoding RNAs in general is presented.
Sizing Up Cellular Noncoding RNAs
Noncoding RNAs populate virtually all polymer lengths between 20 and 20,000 nucleotides and are found in viruses, prokaryotic cells, and eukaryotic cells (Table 1). After they become transcribed, these molecules can be cleaved and chemically modified. As many as 100 potentially functionally meaningful nucleoside modifications have been reported, including nucleobase and sugar methylation, the substitution of sulfur for oxygen, C-glycosidic linkages like the one in pseudouridine, and even condensation products of amino acids and nucleobase exocyclic amino groups (1-3). However, most types of RNA, with the notable exception of transfer RNA (tRNA), are not heavily modified. In keeping with such complexity in primary structure, RNA is known to adopt an impressive array of secondary and tertiary structures, and many of these have been elucidated by molecular modeling, biochemical structure determination, and spectroscopic and crystallographic techniques. Two common RNA secondary structure motifs are the helix, defined as two or more consecutive GC, AU, or GU (canonical) pairs, and the loop, defined as a region in which the nucleotides do not form canonical pairs. Loop structures are further categorized as hairpin, bulge, internal, multibranch, or pseudoknot (4). Beyond Watson-Crick base pairing, ribonucleotides also interact with one another by means of their Hoogsteen and sugar-edge surfaces, and combinations of such pairs as these define specific RNA motifs (5). RNA motifs mediate not only tertiary RNA-RNA contacts, but also quarternary RNA-protein interactions. It should be noted here that, without exception, RNAs within the cell are associated with proteins; however, in the interest of brevity, the discussion below will be limited primarily to the RNA component of each ribonucleoprotein.
Table 1. Some characteristics of noncoding RNAs within the cella
|
RNA* |
Sizec |
Organism |
Physiological role |
|
hTR |
150-1300 nt |
eucarya |
telomerase-mediated telomere synthesis |
|
M1/H1 |
350-450 nt |
universal |
ribonuclease P-mediated pre-tRNA cleavage |
|
group I introns |
200-10000 nt |
bacteria, eucarya0 |
catalyze self-excision from pre-mRNAs, tRNAs, and rRNAs |
|
group II introns |
600-10000 nt |
bacteria, eucarya0 |
catalyze self-excision from pre-mRNAs, tRNAs, and rRNAs, and reverse splicing into DNA |
|
hepatitis 8 ribozyme |
85 nt |
hepatitis δ virus |
catalyzes self-cleavage within viral genomic RNA during rolling-circle replication |
|
hairpin ribozyme |
50 nt |
tobacco ringspot virus |
catalyzes self-cleavage and ligation within viral genomic RNA during rolling-circle replication |
|
hammerhead ribozyme |
78 nt |
S. mansoni |
catalyzes self-cleavage and ligation within viral genomic RNA during rolling-circle replication |
|
snRNA |
100-200 nt |
eucarya |
spliceosome-mediated pre-mRNA splicing |
|
snoRNA |
60-300 nt |
archaea, eucarya |
pre-rRNA processing and modification; snRNA chemical modification |
|
gRNA |
70 nt |
trypanosome protists |
uridine deletion and insertion within mRNA |
|
rRNA0 |
120 nt-4.7 kb |
universal |
catalyzes peptide bond formation in protein biosynthesis |
|
tRNA0 |
70-90 nt |
universal |
protein biosynthesis; transcriptional regulation of aminoacyl-tRNA synthetases |
|
tmRNA |
363 nt |
bacteria |
rescues stalled ribosomes |
|
4.5 S/7 S |
79-522 nt |
universal |
signal recognition particle-mediated protein secretion |
|
siRNA |
20-23 nt |
eucarya |
mRNA cleavage |
|
miRNA |
20-23 nt |
eucarya |
translational repression; mRNA cleavage |
|
rasiRNA |
23-28 nt |
eucarya |
transcriptional silencing; regulation of chromatin structure |
|
tncRNA |
19-23 nt |
C. elegans |
unknown |
|
piRNA |
26-31 nt |
mammals |
suggested antagonists of Piwi proteins |
|
cis-acting RNA (riboswitches) |
100-400 nt |
bacteria0 |
transcriptional and translational regulation of gene expression; respond to temperature, metabolite, protein, or RNA |
|
sRNA |
70-250 nt |
bacteria |
mRNA accumulation, translational activation, mRNA degradation, translational repression |
|
Xist |
17-19 kb |
mammals |
initiates silencing on appropriate X-chromosome |
|
Tsix |
30 kb |
mammals |
blocks initiation of silencing by Xist on appropriate X-chromosome |
|
Xite |
12 kb |
mammals |
mediates persistence of Tsix throughout X-chromosome inactivation |
0See text for references.
0RNA component of ribonucleoprotein complex; in order of appearance in the text.
0Approximate.
0Specifically, in the organelles of eukaryotic cells.
0Although biologists are used to thinking about tRNA, rRNA, and noncoding RNA as altogether different classes of RNA, for historical reasons, tRNA and rRNA are named here as types of noncoding RNA because they do not contain open reading frames that encode proteins.
0Isolated examples of cis-acting elements in eukaryotic mRNAs have been reported, but this type of regulatory mechanism appears to be broadly used only by bacteria.
0Excluding internal open reading frames.
Telomerase is a ribonucleoprotein complex that exists in eukaryotic cells for the apparently sole purpose of synthesizing telomeric DNA, which consists of tandemly repeated sequences that contain clusters of G-residues and forms the ends of chromosomes. Telomerase comprises two essential core components, a protein subunit that has reverse transcriptase (RT) activity and an RNA sequence (hTR) that contains clusters of C-residues and serves as the template substrate for the RT (6). The G-rich DNA and C-rich RNA anneal to form a partial duplex with DNA as the primer. RT-mediated polymerization of dGTP and other complementary triphosphate substrates produces a DNA terminus that has been extended by around six nucleotides. The new end can become a substrate for either another round of telomerase-mediated elongation or primase/polymerase-mediated lagging-strand synthesis.
Processing of RNA
RNA molecules that catalyze RNA strand scission and RNA ligation form one class of ribozymes, or RNA-based enzymes. Well-characterized examples of naturally occurring ribozymes include the RNA moiety of ribonuclease P (M1 in bacteria, H1 in mammals) (7), group I introns (8), group II introns (9, 10), and the hepatitis delta virus (11), hairpin (12), and hammerhead ribozymes (13). Other classes of noncoding RNA that either guide or participate directly in the cleavage, ligation, or chemical modification of various RNA molecules have emerged more recently. Small nuclear RNA (snRNA) is found in the nucleus of eukaryotic cells and is a component of the spliceosome, which, in conjunction with Sm proteins, processes pre-mRNA and prepares it for export to the cytoplasm (14). Small nucleolar RNA (snoRNA), comprising many distinct individuals, and small Cajal body-specific RNA (scaRNA) are also responsible for processing RNA in eukaryotic cells. snoRNAs reside in the nucleolus and guide the cleavage and chemical modification, including sugar methylation and pseudouridylation, reactions that are necessary for large and small ribosomal RNA subunit assembly and export to the cytoplasm (15). Certain snRNAs are similarly acted upon by snoRNAs (16). scaRNAs are localized to foci in the nucleoplasm, known as Cajal bodies, and are involved in the chemical modification of most snRNAs that are transcribed by RNA polymerase II (17). Finally, the guide RNAs (gRNAs) that are found in trypanosome protists direct the site-specific insertion and deletion of uridines in mRNA sequences, a phenomenon referred to as editing (18).
Synthesis and Trafficking of Protein
Ribosomal RNA (rRNA) was long thought to play a passive, structural role in protein synthesis, but it is now known to also be a ribozyme that catalyzes peptide bond formation (19). The other form of noncoding RNA that participates directly in protein synthesis is tRNA, the so-called adaptor molecule found in all organisms that contains both the anticodon RNA sequence and its cognate amino acid residue as a chemically activated aminoacyl ester (20). Two housekeeping RNAs, tmRNA (21) and 4.5 S RNA (22), a component of the signal recognition particle, function to maintain the competence of the ribosome and to ensure the secretion of certain proteins, respectively.
RNA Interference
RNA interference, a mechanism of gene silencing in eukaryotic cells that relies on very short sequences of single-stranded RNA, is arguably the one area of recent noncoding RNA research that has come most to fruition. Small interfering RNAs (siRNAs), microRNAs (miRNAs), repeat-associated small interfering RNAs (rasiRNAs), and tiny-noncoding RNAs (tncRNAs) are short RNA molecules, approximately 22 nucleotides, that have been best characterized in the nematode, C. elegans (23, 24), but also studied in mammals, fish, insects, and plants. The genes that encode siRNAs and miRNAs are transcribed into double-stranded (dsRNA) or short hairpin primary (pri-miRNA) RNAs, respectively. Whereas dsRNA is exported directly to the cytoplasm, pri-miRNA is initially processed to pre-miRNA by the RNase III-like endonuclease, Drosha, in the nucleus. In the cytoplasm, dsRNA and pre-miRNA are acted upon by the RNase III-like endonuclease, Dicer, to yield short dsRNA molecules with 2-nucleotide 3' overhangs and 5' phosphates that are known as siRNAs. In both cases, one strand of siRNA goes on to form a complex with a member of the Argonaute family of proteins, where it acts as a guide in binding to the mRNA target. This ribonucleoprotein complex is referred to as RISC in the siRNA pathway and miRNP in the miRNA pathway. The single-stranded siRNA then binds to its mRNA target by means of perfect (siRNA) or partial (miRNA) complementarity, and it brings about translational repression that may or may not be accompanied by protein-mediated endonucleolytic cleavage of the mRNA target. The newest class of these small mammalian RNAs, Piwi-interacting RNAs (piRNAs), was named for the interaction of its members with Piwi proteins, a subgroup of the Argonaute family (25, 26). Piwi proteins play a role in cell differentiation, and preliminary evidence suggests that piRNAs act as antagonists of their functions.
Riboswitches
RNA sequences that tightly and selectively bind to a molecule of choice, small or large, can be engineered with relative ease; they are referred to as aptamers. For example, in vitro selections have enabled the isolation of RNA sequences that bind to biologically interesting molecules like nucleotides (27), coenzymes (28), amino acids (29), sugars (30), and many others (31). In light of this observation, it is perhaps not surprising that this intrinsic property of RNA is also exploited by nature for the purpose of regulating gene expression with the utmost economy. Riboswitches are cis-acting regulatory RNAs that are found in the 5' untranslated region (5' UTR) of some mRNA molecules and are able to modulate gene expression in response to small molecule metabolites by means of their aptamer and expression platform domains (32). The structures of the domains are coupled in such a way that binding of the ligand directly changes the efficiency of transcription or translation of the mRNA, allowing the cell to circumvent the need for a protein-based metabolite sensor (33). Metabolite-responsive riboswitches represent one type of cis-acting regulatory RNA. Other types respond to temperature, protein ligands, and RNA ligands. Although these types will not be further discussed, recent reports regarding their structure and function are recommended to the reader (34-38).
The nature of the expression platform domain of the riboswitch, and the way in which its structure is coupled to the aptamer domain, in the presence and absence of ligand, dictates whether the riboswitch will affect the extent of transcription or the extent of translation of the mRNA of which it is part (39). Illustrative examples are given below for both transcription-based and translation-based mechanisms. Other less general riboswitch strategies have been reported, including RNA splicing (40, 41), self-cleavage (30), antisense (42), a potential role for RNase P (43), and ribosome-mediated attenuation (44), but these strategies will not be discussed further.
Transcription Effects
Genes are encoded by DNA and become transcribed into RNA through the work of RNA polymerases. This process has three distinct phases: initiation, elongation, and termination (45). On average, elongation in bacteria is highly processive, with about 50 nucleotides per second being incorporated over the length of the transcription unit; however, there do exist natural pause sites at which the rate of transcription slows even though the elongating complex does not dissociate. Protein factors also exist, like NusA, that act to couple the transcription and translation of protein-coding genes by specifically modulating the rate of elongation at specific sites along the DNA template. This phenomenon is exemplified by amino acid biosynthetic operons, most of which contain attenuator sites. In contrast, the termination event is characterized both by an abrupt change of conformation of the elongation complex to an open form and by the dissociation of the completed transcript. Transcription termination occurs when the RNA polymerase encounters either an intrinsic or a Rho-dependent termination sequence within the transcription unit. The kinetic and thermodynamic aspects of transcription elongation and termination, although outside the scope of this review, are important for understanding how the following riboswitches work.
It is during the elongation phase of transcription that many riboswitches achieve either positive or negative regulation of gene expression. After transcription has been initiated and the transcription elongation complex (TEC) has synthesized some portion of the RNA transcript, conserved sequence elements that constitute the aptamer domain of the riboswitch selectively bind to the cognate ligand. The binding event results in the formation of either of two structures in the expression platform domain of the riboswitch: an antiterminator, through which the TEC can proceed without dissociating, or a terminator, through which the TEC cannot proceed without dissociating (Fig. 1). It was recently shown that the kinetics of ligand binding, elongation, and RNA secondary structure formation are of paramount importance to the “decision” to transcribe or not to transcribe, and this may prove to be the case generally (46). Thus, if binding of the ligand results in the formation of an antiterminator, transcription of the complete gene or operon will occur, which can be thought of as positive regulation by a riboswitch. Alternatively, negative regulation by a riboswitch occurs when the ligand-binding event results in the formation of a terminator, precluding transcription of the downstream protein-coding genes.
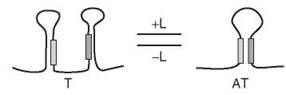
Figure 1. Representation of transcriptional regulation by the binding of a ligand to a c/s-regulatory element of nascent mRNA. Depicted is positive regulation: In the presence of ligand (L), the RNA segment favors a conformation in which an antiterminator (AT), rather than a terminator (T), is present and polymerase-mediated transcription proceeds. In the case of negative regulation, which is not depicted, the presence of ligand (L) causes the RNA segment to favor a conformation in which a terminator (T), rather than an antiterminator (AT), is present, halting polymerase-mediated transcription. The filled bars serve only as orientation guides and do not imply a certain extent of complementarity or stability within the hairpins.
Two examples of positive regulation by a riboswitch in B. subtilis are the glycine-responsive glycine cleavage system (gcvT operon) and the adenine-responsive putative adenine efflux pump (ydhL). In the cell, glycine is important both for its role in protein synthesis as an amino acid and for its role in energy production as a carbon source in the citric acid cycle. Adenine is also of use to the cell, but its relatively low solubility in the cytoplasm creates a need for its removal when it is present in excess. Although the metabolite-binding event brings about antiterminator formation in both the gcvT and the ydhL riboswitches, enabling transcription of the respective downstream genes, the mechanistic details differ significantly between the two regulatory RNAs. Similar to the vast majority of known riboswitches, the ydhL riboswitch has one aptamer domain that binds one ligand molecule (47). In contrast, the gcvT riboswitch contains two aptamer domains that cooperatively bind one glycine molecule each (32). Both aptamer domains have phylogenetically conserved core sequences, and the intervening linker domain shows some conservation in sequence and length. The gcvT riboswitch is only competent when it is in a fully bound state and, importantly glycine binding is cooperative: the binding affinity of the second glycine is 100- to 1000-fold greater than that of the first glycine. This cooperativity of ligand binding, common in protein molecules, has the effect of increasing the sensitivity of the gcvT riboswitch, allowing dramatic changes in transcription to take place in response to small changes in the concentration of glycine. It was suggested that this mechanism, found in the same study to also exist in the putative sodium and alanine symporter encoded by the VC1422 gene of V. cholerae, evolved to ensure that excess glycine is efficiently used as an energy source.
Riboswitches negatively regulate gene expression if binding of the metabolite to the aptamer domain results in the formation of a terminator in the expression platform. Examples of riboswitches that conform to this mechanism are more numerous, as are associated structural studies. High resolution crystal structures exist for the liganded forms of both the 68 nucleotide aptamer domain of the guanine-sensing xpt riboswitch of B. subtilis and an S-adenosylmethionine (SAM)-sensing riboswitch from T. tengcongensis (48, 49). In the G-riboswitch structure, the ligand is stacked within a five-tiered arrangement of base triples, and it participates in eight direct H-bonding interactions with four aptamer nucleotides: three H-bonds represent a canonical Watson-Crick base pair between guanine and a C nucleotide, determining specificity of the riboswitch; three H-bonds are formed between the minor groove heteroatoms of guanine and the Watson-Crick base edge of a U nucleotide; the two remaining H-bonds involve the minor groove oxygen and 2' OH of two independent U nucleotides. In the SAM-riboswitch structure, the ligand adopts a compact conformation that enables both the formation of a cation-pi interaction between its main chain amino group and adenine moiety, and the possibility of participating in two distinct sets of aptamer interactions: the side of the ligand that presents the Watson-Crick face of adenine and the main chain atoms of methionine is engaged in H-bonding and stacking interactions with one helix while the ribose ring and side chain of SAM have favorable van der Waals interactions with a different helix. Despite their differences in ligand binding, the two riboswitches share a remarkably similar overall structure in which a key helix, the P1 helix, is formed as part of either a three-helix tuning-fork-type structure (G-riboswitch) or a four-way junction structure (SAM-riboswitch). In the presence of the cognate metabolite, each structure stabilizes an adjacent terminator helix and prevents transcription of the downstream genes. In the absence of the cognate metabolite, the P1 helix and the terminator dissociate, allowing the nucleotides that otherwise form the 3' side of the P1 helix to form an antiterminator helix with the nucleotides that otherwise form the 5' side of the terminator. This mechanism is thought to be shared by other riboswitches that operate at the level of transcription, such as the FMN-responsive riboswitch that negatively regulates expression of the ribDEAHT operon in B. subtilis (50) and the divalent magnesium-sensing riboswitch that negatively regulates mgtA expression in S. Typhimurium (51).
Translation Effects
Protein biosynthesis in prokaryotes also proceeds through distinct phases known as initiation, elongation, and termination, which are not to be confused with the phases of transcription (see above). Initiation is characterized by the assembly of the small subunit (30 S, comprising 21 proteins and the 1500 nucleotide 16 S RNA) and the large subunit (50 S, composed of 34 proteins, the 120 nucleotide 5S RNA, and the 2900 nucleotide 23 S RNA) of the ribosome, together with the initiator tRNA that is charged with N-formylmethionine, on the 30 nucleotide ribosome-binding site (RBS) of a given mRNA (52). The RBS, located within the 5' UTR of the transcript, has several important features that are common among most mRNAs. From 5' to 3', there is a pyrimidine-rich region that interacts with ribosomal protein S1; the Shine-Dalgarno (SD) sequence, typically GGAGG, forms base pairs with the 16 S rRNA; the initiation codon is AUG in the majority of cases, and GUG or UUG in a few cases; finally, there is a downstream box (DB) that also forms base pairs with the 16 S rRNA. Therefore, the precise sequence and structure of the mRNA are of paramount importance to the efficiency with which it becomes translated into protein. Many riboswitches regulate gene expression at the stage of translation initiation. Their mechanisms are analogous to riboswitch regulation at the transcriptional level, described above, except that ligand binding results in the formation of a structure that either sequesters or liberates the RBS.
Atomic resolution structures of the liganded forms of both the 71 nucleotide adenine-sensing aptamer domain of the add riboswitch in V. vulnificus, an example of positive translational regulation, and the 80 nucleotide thiamine pyrophosphate (TPP)-sensing aptamer domain of the thiM riboswitch in E. coli , an example of negative translational regulation, were recently solved. The structure of the A-riboswitch is very similar to that of the G-riboswitch, described above, except that the specificity-determining pyrimidine is a U rather than a C, and that the tuning fork structure containing the P1 helix coexists with a single-stranded downstream region in which the RBS is available to bind the 30 S subunit of the ribosome (48). In this case, the expression of adenine deaminase is increased in the presence of adenine. In contrast, translation of thiM mRNA is reduced in the presence of TPP, and this negative regulatory mechanism is also made clear by inspection of the X-ray crystal structure (53). In the complex, TPP adopts an extended conformation in which it acts as a bridge between two parallel helical domains that are connected to the P1 helix by means of a three-way junction. The 4-amino-5-hydroxymethyl-2-methylpyrimidine moiety of the ligand engages in intercalation, H-bonding, base stacking, and base triple formation with one helical domain while the pyrophosphate group is bound to the other helical domain by H-bonding and electrostatic interactions that are mediated by two hexacoordinated divalent magnesium ions. The P1 helix stabilizes a downstream hairpin in which the SD sequence and start codon of thiM are sequestered, precluding translation initiation. It is likely that similar structures play a role in the negative translational regulation of ypaA in B. subtilis by a flavin mononucleotide (FMN)-responsive riboswitch (50), and that of btuB in E. coli by a coenzyme B12-responsive riboswitch (54).
Chromosomally Encoded Regulatory sRNAs in Bacteria
sRNAs that regulate gene expression in bacteria are a newly identified class of noncoding RNAs. These molecules are commonly 70-200 nucleotides in length and are found encoded within the chromosome in either a cis (antisense) or a trans location relative to their target protein-coding genes. Although sRNAs can influence gene expression by interacting with protein targets, most of the individuals that have been characterized thus far act by means of either extensive or limited base pairing with mRNA targets. This class of regulators was discovered long ago in the plasmids harbored by enterobacteria, in bacteriophage, and in transposons; all of these are cis -encoded antisense RNAs that exploit their perfect complementarity to bind to the cognate transcript, which encodes a replication factor, toxin, or transposase, to name a few, and bring about transcription attenuation, mRNA degradation, or inhibition of translation (55). Within the past 5 years, much more extensive work has been done to identify and characterize sRNAs that are encoded within the genomes of bacteria. The history of our knowledge of such sRNAs has been described as a chronology with two distinct periods: the classic and the modern (56). In the classic age, about 12 novel RNA molecules were discovered, either intentionally by means of metabolic labeling or serendipitously. In the modern age, which began in 2001, the existence of new information, generated by new technology, prompted eight research groups to carry out comprehensive, systematic searches for more sRNAs (57-64). Both computational methods and direct experimental detection were used, and these contributions increased the number of confirmed chromosomally encoded sRNAs in E. coli to about 70 (65). Although characterization of the physiological roles and mechanisms of action of these sRNAs is still in its early stages, the following summary of the work done so far illustrates that their expression patterns and functions are too diverse to be generalized.
Regulation of sRNAs
It is known that the steady-state level of many sRNAs depends on the phase of growth or environmental conditions of the cell, and obviously the concentration of the sRNA at any given time represents a sum of its expression, processing, and degradation. With respect to expression, some regulatory mechanisms have been solved: RprA sRNA is positively regulated by the RcsC/RcsB phosphorelay system (66); OxyS sRNA is induced in the presence of hydrogen peroxide by the transcriptional regulator, OxyR (67); IstR-2 sRNA is encoded adjacent to a LexA protein-binding site, and its expression is induced by treating cells with the DNA-damaging agent, mitomycin C (68); RyhB sRNA is encoded adjacent to a Fur protein-binding site, and its expression is induced in response to iron depletion (69); OmrA (RygA), OmrB (RygB), and MicF sRNAs are regulated in part by the EnvZ-OmpR two-component system, which responds to changes in osmolarity (70). Apparently, many sRNAs are present in the cell in more than one length; for example, GadY was shown to exist simultaneously as 105, 90, and 59 nucleotide RNA molecules (71). However, the details of sRNA biogenesis regarding active and inactive forms and the existence or extent of chemical modification are not well understood. Finally, the half-lives of sRNAs differ significantly from one to the next (61). As the protein factors that are known to play a role in the degradation of some sRNAs tend to be the same as those that are involved in the sRNA-mediated regulation of gene expression, they are described in the following section.
Regulation by sRNAs: RNA-RNA Interaction
Posttranscriptional regulation of gene expression can be achieved by base pairing interactions between sRNAs and their target mRNAs. It is important to note that the two sequences need not be perfectly complementary over a long patch; in fact, relatively short regions of complementarity interrupted by mismatches, small loops, or longer intervening sequences are the norm. The sRNA-mRNA binding event has been shown to result in a variety of outcomes, including mRNA accumulation, activation of translation, repression of translation, and mRNA degradation. Thus far, the involvement in these processes of three main proteins has been demonstrated, although the extent and nature of their roles cannot be generalized. First, the Sm-like host factor for QP replicase (Hfq) is an RNA-binding protein. It has been shown to have a high binding affinity for more than one third of the 70 known sRNAs, and to increase the half-lives and steady-state levels of certain sRNAs (72). Moreover, multiple studies have shown that it enhances interaction between sRNAs and their target mRNAs (73-75). Structural studies of Hfq have contributed to the development of a model in which the protein binds to relatively structureless A/U-rich segments of an RNA molecule that are in the vicinity of highly structured regions, and that the binding event has long-range effects on these secondary structures that allow intermolecular RNA-RNA interactions to take place (73, 76). Second, RNase E is a hydrolytic endonuclease that cleaves single-stranded RNA (77). It has been shown to catalyze the degradation of certain sRNAs and their target mRNAs, some examples of which are given below. Whereas its N-terminal catalytic domain adopts a compact fold and is well-conserved across species, the C-terminal domain of RNase E is neither generally structured nor conserved, consisting rather of 15-40 amino acid stretches, referred to as microdomains, that bind to known protein partners like RhlB, enolase, and PNPase, components of the degradosome, but may also interact with other protein and RNA factors or enable self-assembly (78). For example, Hfq itself has been shown to interact with the C-terminus of RNase E (79). Third, RNase III is a hydrolytic endonuclease that cleaves double-stranded RNA (80). It has also been shown to catalyze the degradation of certain sRNAs and their target mRNAs, some examples of which are given below.
An sRNA can increase expression of its target gene, an outcome that appears to be relatively rare, by increasing the amount of target mRNA, increasing the efficiency with which it becomes translated into protein, or doing both. The cis-encoded antisense sRNA, GadY, was recently shown to increase the level of gadX mRNA in E. coli by means of perfectly complementary base pairing interactions involving the 3' UTRs of both RNAs (71). The increase in gadX mRNA, which encodes a transcriptional regulator of acid resistance genes, eventually results in an increase in the levels of two glutamate decarboxylases, GadA and GadB. Examples of sRNAs that make use of a translational activation mechanism are also few; however, DsrA (85 nucleotides) and RprA (105 nucleotides) are two sRNAs that act to positively and nonredundantly regulate the translation of rpoS mRNA into its product, the stationary phase sigma transcription factor, in E. coli (66, 81). In the absence of DsrA and RprA, the intramolecular hairpin structure adopted by the unusually long (567 nucleotide) 5' UTR of rpoS mRNA sequesters its RBS, precluding translation initiation. When present, DsrA and RprA form base pairs with the upstream region of the 5' UTR of rpoS mRNA, competing with interactions it otherwise engages in with the downstream region of the 5' UTR and liberating the RBS. Both DsrA-mediated and RprA-mediated translational regulation of rpoS are Hfq-dependent (82).
In most cases described to date, base pairing of an sRNA to its target mRNA results in a negative effect on gene expression. By analogy to positive sRNA-mediated gene regulation, the RNA regulator can achieve its negative effect by decreasing the amount of target mRNA, decreasing the efficiency with which it becomes translated into protein, or doing both. The strategy in which an sRNA represses translation by directly binding to the RBS of its target mRNA, in an Hfq-dependent manner, is highly represented in the regulation of expression of outer membrane porins (Omp) in E. coli: ompC, ompF, and ompA are selectively regulated in this way by MicC, MicF, and MicA sRNAs, respectively (83-86). Other sRNAs have been shown to bring about target mRNA cleavage by means of Hfq and RNase E. For example, RyhB sRNA leads to the degradation of sodB (encodes iron-containing superoxide dismutase) and sdh (encodes iron-containing succinate dehydrogenase) mRNAs under iron-depleted conditions, and SgrS sRNA leads to the degradation of ptsG mRNA, which encodes a major glucose transporter, under sugar phosphate stress (69, 87, 88). Two important aspects of this mechanism have recently come to light. First, the C-terminal domain of RNase E interacts with Hfq, and a ribonucleoprotein complex composed of these two proteins and an sRNA is sufficient to bring about specific mRNA degradation (79). Second, although target cleavage often accompanies sRNA-mediated translational repression, making the effect irreversible, it may not be necessary to achieve the desired regulatory effect (89). In some cases, degradation of target mRNA in the presence of an sRNA has been shown to depend on RNase III rather than RNase E. For example, IstR-1 sRNA and tisAB mRNA, which encodes a toxic peptide that is induced as part of the SOS response in E. coli, are trans-encoded but remarkably share a continuous stretch of 23 canonical base pairs of complementarity. IstR-1 and tisAB undergo cleavage in vivo only in the presence of one another and RNase III, and IstR-1 is necessary for suppressing the toxic effects of the TisB peptide under the conditions (SOS-on) in which it becomes transcribed (68). In a separate study, it was suggested that a similar mechanism could account for the decrease in the number of IsiA-photosystem I supercomplexes caused by overexpression of the 177 nucleotide antisense IsrR sRNA, which is cis -encoded with respect to its target, isiA, in the cyanobacterium, Synechocystis sp. PCC 6803 (90). The common feature shared by the IstR-1/tisAB and IsrR/isiA regulatory systems is a high degree of complementarity between the sRNA and its target mRNA, which likely enables the formation of a double-stranded RNA structure that is recognized as a substrate by RNase III.
Regulation by sRNAs: RNA-Protein Interaction
Although the majority of chromosomally encoded regulatory sRNAs in bacteria, whose functions have been described, work by base pairing directly to an RNA target, there are also cases in which sRNAs have been shown to bind to a protein target to change the expression of genes at either the transcriptional or the posttranscriptional level. For example, 6 S sRNA is known to impact global partitioning of transcription by directly interacting with a transcription factor. After becoming transcribed, together with an open reading frame as a discistronic unit, and processed, to a 184 nucleotide sRNA, 6S goes on to tightly and specifically bind RpoD protein (91). RpoD, also known as σ70, is the vegetative transcription factor that directs RNA polymerase to transcribe the subset of genes that ought to be expressed under normal conditions. By folding into a long hairpin with a central internal loop that very much resembles an open promoter of DNA, 6 S RNA is thought to compete with actual promoter DNA for the nucleic acid-binding site of RpoD, which in turn allows other regulons that use alternative sigma factors to become more expressed. A further testament to the versatility of sRNAs in bacteria is the paradigmatic CsrA protein/CsrB RNA system of posttranscriptional regulation. CsrA is a posttranscriptional regulator that binds to mRNAs, which code for a diverse set of proteins but share a conserved core sequence that the protein recognizes. By changing the stability or efficiency of translation of its targets, CsrA can have either positive or negative effects on their expression (92). CsrB and CsrC are sRNAs, 360 and 245 nucleotides in length, respectively, that bind CsrA by means of repeated hairpins that terminate in loops composed of the same conserved core sequence found in the mRNA targets of CsrA (93, 94). Whereas CsrB and CsrC comprise 18 and 9 such loops, respectively, the mRNA targets of CsrA have only one to three per molecule. In binding CsrA, CsrB and CsrC antagonize the numerous and diverse effects that this protein otherwise has as a posttranscriptional regulator.
Large Noncoding RNAs and Mammalian X-Chromosome Inactivation
X-chromosome inactivation (XCI) is the process by which female mammalian cells, early in development, silence nearly all of the genes on all but one X-chromosome to achieve dosage parity between the sexes (95). Four distinct processes are associated with XCI: X-chromosome counting, chromosome choice, initiation and propagation of silencing, and maintenance of heterochromatin. All of these events are regulated by an 80-450-kb X-linked locus called the X-inactivation center (Xic). Interestingly, this locus can be described as having a dearth of protein-coding sequences and an abundance of genes that encode large noncoding RNAs. As the detailed mechanisms behind large noncoding RNA involvement in XCI have not yet been solved, it is important to keep in mind the full range of potential models that have been proposed (96). First, by virtue of its DNA sequence, a gene itself is able to bind proteins, transcription factors for example, thereby affecting the extent of transcription of other genes that are either proximal or distal. Second, when a eukaryotic gene is transcribed, local changes in chromatin structure occur and can affect the transcription of other genes in the neighborhood; such regulation is also known to occur in short (SINEs) and long interspersed nucleotide elements (LINEs) (97). Third, the RNA product could recruit suppressive factors or form duplex RNA, leading to either degradation of the nucleic acid target or its inability to associate with the necessary protein factors.
The first large noncoding RNA gene of the Xic to be identified was Xist (98). In mice, it encodes a 17.4 kb untranslated RNA that is essential for XCI. Xist is expressed from the inactivated X-chromosome and interacts physically with it, in cis, via chromatin and the nuclear matrix, “coating” the chromosome. According to the current model, two conserved sequences in Xist, Repeat A and Repeat C, affect transcription from both the Xist promoter and promoters subject to XCI and recruit specific silencing proteins to the Xic, respectively. Likely protein partners in this process are enzymes that methylate or ubiquitylate chromatin. As Xist is transcribed, a process that could last up to 30 minutes, it spreads along the silenced chromosome and is thought to propagate these transcriptional and protein-recruiting effects as it goes. Xist is repressed by the antisense large noncoding RNA, Tsix (98). Before silencing, Tsix and Xist are both expressed, with Tsix being present in excess, from both the X-chromosome to remain active and the X-chromosome to be silenced. Once chromosomal choice has occurred, Tsix is only expressed on the active X-chromosome until the process of XCI has ended. Tsix is encoded antisense to Xist, its transcription start site is downstream of Xist, and its function requires that it be transcribed to at least within the Xist gene. The persistence of Tsix is aided by another large noncoding RNA, Xite (X-inactivation intergenic transcription elements), which has multiple start sites clustered in two regions upstream of Tsix (98).
Although it is not yet known exactly how these and other large noncoding RNAs help to regulate XCI, and the related process of genomic imprinting, their mechanisms are likely to be complex. It is interesting to speculate that this particular physiological role is ideally suited to RNAs rather than proteins. That is, because protein-coding RNAs are transported to the cytoplasm for translation, their protein products are not usually traceable to the chromosome of origin of their corresponding transcript. Differently, noncoding RNAs remain in the nucleus to achieve the ends for which they evolved.
References
1. Limbach PA, Crain PF, McClosky JA. Summary: the modified nucleosides of RNA. Nucleic Acids Res. 1994; 22(12): 2183-2196.
2. Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X. Methylation as a crucial step in plant microRNA biogenesis. Science 2005; 307(5711): 932-935.
3. Ebhardt HA, Thi EP, Wang MB, Unrau PJ. Extensive 3; modification of plant small RNAs is modulated by helper component-proteinase expression. Proc. Natl. Acad. Sci. U.S.A. 2005; 102(38): 13398-13403.
4. Ruschak AM, Mathews DH, Bibillo A, Spinelli SL, Childs JL, Eickbush TH, Turner DH. Secondary structure models of the 3; untranslated regions of diverse R2 RNAs. RNA 2004; 10(6): 978-987.
5. Leontis NB, Westhof E. Analysis of RNA motifs. Curr. Opin. Struct. Biol. 2003; 13(3): 300-308.
6. Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 1985; 43(2 Pt 1): 405-413.
7. Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 1983; 35(3 Pt 2): 849-857.
8. Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, Cech TR. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell 1982; 31(1): 147-157.
9. Lehmann K, Schmidt U. Group II introns: structure and catalytic versatility of large natural ribozymes. Crit. Rev. Biochem. Mol. Biol. 2003; 38(3): 249-303.
10. Lambowitz AM, Zimmerly S. Mobile group II introns. Annu. Rev. Genet. 2004; 38: 1-35.
11. Shih IH, Been MD. Catalytic strategies of the hepatitis delta virus ribozymes. Annu. Rev. Biochem. 2002; 71: 887-917.
12. Ferre-D’Amare AR. The hairpin ribozyme. Biopolymers 2004; 73(1): 71-78.
13. Blount KF, Uhlenbeck OC. The hammerhead ribozyme. Biochem. Soc. Trans. 2002; 30(Pt 6): 1119-1122.
14. Patel AA, Steitz JA. Splicing double: insights from the second spliceosome. Nat. Rev. Mol. Cell Biol. 2003; 4(12): 960-970.
15. Granneman S, Baserga SJ. Ribosome biogenesis: of knobs and RNA processing. Exp. Cell Res. 2004; 296(1): 43-50.
16. Tycowski KT, You ZH, Graham PJ, Steitz JA. Modification of U6 spliceosomal RNA is guided by other small RNAs. Mol. Cell. 1998; 2(5): 629-638.
17. Darzacq X, Jady BE, Verheggen C, Kiss AM, Bertrand E, Kiss T. Cajal body-specific small nuclear RNAs: a novel class of 2-O-methylation and pseudouridylation guide RNAs. EMBO J. 2002; 21(11): 2746-2756.
18. Blum B, Bakalara N, Simpson L. A model for RNA editing in kinetoplastid mitochondria: “guide” RNA molecules transcribed from maxicircle DNA provide the edited information. Cell 1990; 60(2): 189-198.
19. Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science 2000; 289(5481): 920-930.
20. Ribas de Pouplana L, Schimmel P. Aminoacyl-tRNA synthetases: potential markers of genetic code development. Trends Biochem. Sci. 2001; 26(10): 591-596.
21. Keiler KC, Waller PR, Sauer RT. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 1996; 271(5251): 990-993.
22. Keenan RJ, Freymann DM, Stroud RM, Walter P. The signal recognition particle. Annu. Rev. Biochem. 2001; 70: 755-775.
23. Filipowicz W. RNAi: the nuts and bolts of the RISC machine. Cell 2005; 122(1): 17-20.
24. Aravin A, Tuschl T. Identification and characterization of small RNAs involved in RNA silencing. FEBS Lett. 2005; 579(26): 5830-5840.
25. Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins.Nature 2006; 442(7099): 199-202.
26. Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, Chien M, Russo JJ, Ju J, Sheridan R, Sander C, Zavolan M, Tuschl T. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 2006; 442(7099): 203-207.
27. Sazani PL, Larralde R, Szostak JW. A small aptamer with strong and specific recognition of the triphosphate of ATP. J. Am. Chem. Soc. 2004; 126(27): 8370-8371.
28. Burgstaller P, Famulok M. Isolation of RNA aptamers for biological cofactors by in-vitro selection. Angew. Chem. Int. Ed. Engl. 1994; 33(10): 1084-1087.
29. Mannironi C, Scerch C, Fruscoloni P, Tocchini-Valentini GP. Molecular recognition of amino acids by RNA aptamers: the evolution into an L-tyrosine binder of a dopamine-binding RNA motif. RNA 2000; 6(4): 520-527.
30. Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. Control of gene expression by a natural metabolite-responsive ribozyme. Nature 2004; 428(6980): 281-286.
31. Lee JF, Hesselberth JR, Meyers LA, Ellington AD. Aptamer database. Nucleic Acids Res. 2004; 32(Database issue): D95-100.
32. Mandal M, Lee M, Barrick JE, Weinberg Z, Emilsson GM, Ruzzo WL, Breaker RR. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science 2004; 306(5694): 275-279.
33. Winkler WC. Metabolic monitoring by bacterial mRNAs. Arch. Microbiol. 2005; 183(3): 151-159.
34. Johansson J, Mandin P, Renzoni A, Chiaruttini C, Springer M, Cossart P. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell 2002; 110(5): 551-561.
35. Chowdhury S, Maris C, Allain FH, Narberhaus F. Molecular basis for temperature sensing by an RNA thermometer. EMBO J. 2006; 25(11): 2487-2497.
36. Otridge J, Gollnick P. MtrB from Bacillus subtilis binds specifically to trp leader RNA in a tryptophan-dependent manner. Proc. Natl. Acad. Sci. U.S.A. 1993; 90(1): 128-132.
37. Antson AA, Dodson EJ, Dodson G, Greaves RB, Chen X, Gollnick P. Structure of the trp RNA-binding attenuation protein, TRAP, bound to RNA. Nature 1999; 401(6750): 235-242.
38. Nelson AR, Henkin TM, Agris PF. tRNA regulation of gene expression: interactions of an mRNA 5’-UTR with a regulatory tRNA. RNA 2006; 12(7): 1254-1261.
39. Winkler WC. Riboswitches and the role of noncoding RNAs in bacterial metabolic control. Curr. Opin. Chem. Biol. 2005; 9(6): 594-602.
40. Kubodera T, Watanabe M, Yoshiuchi K, Yamashita N, Nishimura A, Nakai S, Gomi K, Hanamoto H. Thiamine-regulated gene expression of Aspergillus oryzae thiA requires splicing of the intron containing a riboswitch-like domain in the 5;-UTR. FEBS Lett. 2003; 555(3): 516-520.
41. Sudarsan N, Barrick JE, Breaker RR. Metabolite-binding RNA domains are present in the genes of eukaryotes. RNA 2003; 9(6): 644-647.
42. Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. Comparative genomics of the methionine metabolism in Grampositive bacteria: a variety of regulatory systems. Nucleic Acids Res. 2004; 32(11): 3340-3353.
43. Altman S, Wesolowski D, Guerrier-Takada C, Li Y. RNase P cleaves transient structures in some riboswitches. Proc. Natl. Acad. Sci. U.S.A. 2005; 102(32): 11284-11289.
44. Gollnick P, Babitzke P. Transcription attenuation. Biochim. Bio- phys. Acta. 2002; 1577(2): 240-250.
45. Richardson JP, Greenblatt J. Control of RNA chain elongation and termination. In: Escherichia coli and Salmonella: Cellular and Molecular Biology, 2nd edition. Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, eds. 1996. American Society for Microbiology Press, Washington, D.C., pp. 822-848.
46. Wickiser JK, Winkler WC, Breaker RR, Crothers DM. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol. Cell. 2005; 18(1): 49-60.
47. Mandal M, Breaker RR. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat. Struct. Mol. Biol. 2004; 11(1): 29-35.
48. Serganov A, Yuan YR, Pikovskaya O, Polonskaia A, Malinina L, Phan AT, Hobartner C, Micura R, Breaker RR, Patel DJ. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem. Biol. 2004; 11(12): 1729-1741.
49. Montange RK, Batey RT. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature 2006; 441(7097): 1172-1175.
50. Winkler WC, Cohen-Chalamish S, Breaker RR. An mRNA structure that controls gene expression by binding FMN. Proc. Natl. Acad. Sci. U.S.A. 2002; 99(25): 15908-15913.
51. Cromie MJ, Shi Y, Latifi T, Groisman EA. An RNA sensor for intracellular Mg(2+). Cell 2006; 125(1): 71-84.
52. Laursen BS, Sorensen HP, Mortensen KK, Sperling-Petersen HU. Initiation of protein synthesis in bacteria. Microbiol. Mol. Biol. Rev. 2005; 69(1): 101-123.
53. Serganov A, Polonskaia A, Phan AT, Breaker RR, Patel DJ. Structural basis for gene regulation by a thiamine pyrophosphatesensing riboswitch. Nature 2006; 441(7097): 1167-1171.
54. Nahvi A, Sudarsan N, Ebert MS, Zou X, Brown KL, Breaker RR. Genetic control by a metabolite binding mRNA. Chem. Biol. 2002; 9(9): 1043-1049.
55. Wagner EGH, Altuvia S, Romby P. Antisense RNAs in bacteria and their genetic elements. Adv. Genet. 2002; 46: 361-398.
56. Gottesman S. The small RNA regulators of Escherichia coli: roles and mechanisms. Annu. Rev. Microbiol. 2004; 58: 303-328.
57. Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner EGH, Margalit H, Altuvia S. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 2001; 11(12): 941-950.
58. Wassarman KM, Repoila F, Rosenow C, Storz G, Gottesman S. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 2001; 15(13): 1637-1651.
59. Rivas E, Klein RJ, Jones TA, Eddy SR. Computational identification of noncoding RNAs in E. coli by comparative genomics. Curr. Biol. 2001; 11(17): 1369-1373.
60. Chen S, Lesnik EA, Hall TA, Sampath R, Griffey RH, Ecker DJ, Blyn LB. A bioinformatics based approach to discover small RNA genes in the Escherichia coli genome. Biosystems 2002; 65(2-3): 157-177.
61. Vogel J, Bartels V, Tang TH, Churakov G, Slagter-Jager JG, Huttenhofer A, Wagner EGH. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res. 2003; 31(22): 6435-6443.
62. Kawano M, Reynolds AA, Miranda-Rios J, Storz G. Detection of 5’- and 3’-UTR-derived small RNAs and cis-encoded antisense RNAs in Escherichia coli. Nucleic Acids Res. 2005; 33(3): 1040-1050.
63. Tjaden B, Saxena RM, Stolyar S, Haynor DR, Kolker E, Rosenow C. Transcriptome analysis of Escherichia coli using high-density oligonucleotide probe arrays. Nucleic Acids Res. 2002; 30(17): 3732-3738.
64. Zhang AX, Wassarman KM, Rosenow C, Tjaden BC, Storz G, Gottesman S. Global analysis of small RNA and mRNA targets of Hfq. Mol. Microbiol. 2003; 50(4): 1111-1124.
65. Vogel J, Sharma CM. How to find small non-coding RNAs in bacteria. Biol. Chem. 2005; 386(12): 1219-1238.
66. Majdalani N, Hernandez D, Gottesman S. Regulation and mode of action of the second small RNA activator of RpoS translation, RprA. Mol. Microbiol. 2002; 46(3): 813-826.
67. Altuvia S, Weinstein-Fischer D, Zhang A, Postow L, Storz G. A small, stable RNA induced by oxidative stress: Role as a pleiotropic regulator and antimutator. Cell 1997; 90(1): 43-53.
68. Vogel J, Argaman L, Wagner EG, Altuvia S. The small RNA IstR inhibits synthesis of an SOS-induced toxic peptide. Curr. Biol. 2004; 14(24): 2271-2276.
69. Masse E. Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2002; 99(7): 4620-4625.
70. Guillier M, Gottesman S. Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Mol. Microbiol. 2006; 59(1): 231-247.
71. Opdyke JA, Kang JG, Storz G. GadY, a small-RNA regulator of acid response genes in Escherichia coli. J. Bacteriol. 2004; 186(20): 6698-6705.
72. Valentin-Hansen P, Eriksen M, Udesen C. The bacterial Sm-like protein Hfq: a key player in RNA transactions. Mol. Microbiol. 2004; 51(6): 1525-1533.
73. Moller T, Franch T, Hojrup P, Keene DR, Bachinger HP, Brennan RG, Valentin-Hansen P. Hfq: a bacterial Sm-like protein that mediates RNA-RNA interaction. Mol. Cell. 2002; 9(1): 23-30.
74. Zhang AX, Wassarman KM, Ortega J, Steven AC, Storz G. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol. Cell. 2002; 9(1): 11-22.
75. Kawamoto H, Koide Y, Morita T, Aiba H. Base-pairing requirement for RNA silencing by a bacterial small RNA and acceleration of duplex formation by Hfq. Mol. Microbiol. 2006; 61(4): 1013-1022.
76. Schumacher MA, Pearson RF, Moller T, Valentin-Hansen P, Brennan RG. Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein. EMBO J. 2002; 21(13): 3546-3556.
77. Carpousis AJ. The Escherichia coli RNA degradosome: structure, function and relationship to other ribonucleolytic multienyzme complexes. Biochem. Soc. Trans. 2002; 30: 150-155.
78. Marcaida MJ, DePristo MA, Chandran V, Carpousis AJ, Luisi BF. The RNA degradosome: life in the fast lane of adaptive molecular evolution. Trends Biochem. Sci. 2006; 31(7): 359-365.
79. Morita T, Maki K, Aiba H. RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev. 2005; 19(18): 2176-2186.
80. Sun W, Pertzev A, Nicholson AW. Catalytic mechanism of Escherichia coli ribonuclease III: kinetic and inhibitor evidence for the involvement of two magnesium ions in RNA phosphodiester hydrolysis. Nucleic Acids Res. 2005; 33(3): 807-815.
81. Sledjeski DD, Whitman C, Zhang A. Hfq is necessary for regulation by the untranslated RNA DsrA. J. Bacteriol. 2001; 183(6): 1997-2005.
82. Majdalani N, Chen S, Murrow J, John KS, Gottesman S. Regulation of RpoS by a novel small RNA: the characterization of RprA. Mol. Microbiol. 2001; 39(5): 1382-1394.
83. Chen S, Zhang A, Blyn LB, Storz G. MicC, a second small-RNA regulator of Omp protein expression in Escherichia coli. J. Bacteriol. 2004; 186(20): 6689-6697.
84. Delihas N, Forst S. MicF: an antisense RNA gene involved in response of Escherichia coli to global stress factors. J. Mol. Biol. 2001; 313(1): 1-12.
85. Udekwu KI, Darfeuille F, Vogel J, Reimegard J, Holmqvist E, Wagner EG. Hfq-dependent regulation of OmpA synthesis is mediated by an antisense RNA. Genes Dev. 2005; 19(19): 2355-2366.
86. Rasmussen AA, Eriksen M, Gilany K, Udesen C, Franch T, Petersen C, Valentin-Hansen P. Regulation of ompA mRNA stability: the role of a small regulatory RNA in growth phase-dependent control. Mol. Microbiol. 2005; 58(5): 1421-1429.
87. Masse E, Escorcia FE, Gottesman S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003; 17(19): 2374-2383.
88. Vanderpool CK, Gottesman S. Involvement of a novel transcriptional activator and small RNA in post-transcriptional regulation of the glucose phosphoenolpyruvate phosphotransferase system. Mol. Microbiol. 2004; 54(4): 1076-1089.
89. Morita T, Mochizuki Y, Aiba H. Translational repression is sufficient for gene silencing by bacterial small noncoding RNAs in the absence of mRNA destruction. Proc. Natl. Acad. Sci. U.S.A. 2006; 103(13): 4858-4863.
90. Duhring U, Axmann IM, Hess WR, Wilde A. An internal antisense RNA regulates expression of the photosynthesis gene isiA. Proc. Natl. Acad. Sci. U.S.A. 2006; 103(18): 7054-7058.
91. Wassarman KM, Storz G. 6 S RNA regulates E. coli RNA polymerase activity. Cell 2000; 101(6): 613-623.
92. Romeo T. Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Mol. Microbiol. 1998; 29(6): 1321-1330.
93. Liu MY, Gui G, Wei B, Preston JF III, Oakford L, Yuksel U, Giedroc DP, Romeo T. The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J. Biol. Chem. 1997; 272(28): 17502-17510.
94. Weilbacher T, Suzuki K, Dubey AK, Wang X, Gudapaty S, Morozov I, Baker CS, Georgellis D, Babitzke P, Romeo T. A novel sRNA component of the carbon storage regulatory system of Escherichia coli. Mol. Microbiol. 2003; 48(3): 657-670.
95. Plath K, Mlynarczyk-Evans S, Nusinow DA, Panning B. Xist RNA and the mechanism of X chromosome inactivation. Annu. Rev. Genet. 2002; 36: 233-278.
96. Boumil RM, Lee JT. Forty years of decoding the silence in X-chromosome inactivation. Hum. Mol. Genet. 2001; 10(20): 2225-2232.
97. Druker R, Whitelaw E. Retrotransposon-derived elements in the mammalian genome: a potential source of disease. J. Inherit. Metab. Dis. 2004; 27(3): 319-330.
98. Andersen AA, Panning B. Epigenetic gene regulation by noncoding RNAs. Curr. Opin. Cell Biol. 2003; 15(3): 281-289.
Further Reading
Barciszewski J, Erdmann VA, eds. Noncoding RNAs: Molecular Biology and Molecular Medicine. 2003. Eurekah.com and Kluwer Academic/Plenum Publishers, London, UK.
Gesteland RF, Cech TR, Atkins JF, eds. The RNA World, 3rd edition. 2006. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. See also first and second editions.
Hartmann RK, Bindereif A, Schon A, Westhof E, eds. Handbook of RNA Biochemistry. 2005. Wiley-VCH, Weinheim, Germany.
Cao X, Yeo G, Muotri AR, Kuwabara T, Gage FH. Noncoding RNAs in the mammalian central nervous system. Annu. Rev. Neurosci. 2006; 29: 77-103.
Eddy SR. Non-coding RNA genes and the modern RNA world. Nat. Rev. Genet. 2001; 2(12): 919-929
Gottesman S. Micros for microbes: non-coding regulatory RNAs in bacteria. Trends Genet. 2005; 21(7): 399-404.
Huttenhofer A, Vogel J. Experimental approaches to identify non-coding RNAs. Nucleic Acids Res. 2006; 34(2): 635-646.
Storz G, Altuvia S, Wassarman KM. An abundance of RNA regulators. Annu. Rev. Biochem. 2005; 74: 199-217.
Storz G, Opdyke JA, Zhang A. Controlling mRNA stability and translation with small, noncoding RNAs. Curr. Opin. Microbiol. 2004; 7(2): 140-144.
Szymanski M, Barciszewska MZ, Erdmann VA, Barciszewski J. A new frontier for molecular medicine: noncoding RNAs. Biochim. Biophys. Acta. 2005; 1756(1): 65-75.
The microRevolution. Nat. Genet. 2006; 38: S1-S36.
http://www.prl.msu.edu/PLANTncRNAs/
http://www.sanger.ac.uk/Software/Rfam/
http://research.imb.uq.edu.au/rnadb/
See Also
micro RNA (miRNA), Chemistry of
Ribozymes Selected by Nature, Chemistry of
RNA, small nuclear (snRNA) and small nucleolar (snoRNA), Chemistry of
small interfering RNA (siRNA), chemistry of
mRNA Untranslated Regions (UTRs)