CHEMICAL BIOLOGY
Molecular Chaperones
R. John Ellis, University of Warwick, Coventry, United Kingdom
doi: 10.1002/9780470048672.wecb068
The generic tendency of proteins to aggregate into nonfunctional, and sometimes cytotoxic, structures poses a universal problem for all types of cells. This problem is exacerbated by the high total concentrations of macromolecules found within most intracellular compartments, but it is solved by the actions of certain proteins that function as molecular chaperones. Different chaperones act by distinct mechanisms on both the folding of polypeptide chains and their subsequent assembly into oligomeric structures. Many chaperones, but not all, are also stress (or heat shock) proteins because the need for a chaperone function increases under stress conditions that cause proteins to unfold.
Proteins have evolved over billions of years to function inside highly complex, intracellular environments, but their properties commonly are studied after purification and after exposure to much simpler and very different conditions in the test tube. Christian Anfinsen received the Nobel Prize for chemistry in 1972 for discovering that some purified denatured proteins will refold into their biologically active conformations when the concentration of the denaturing agent is lowered (1). This classic type of refolding experiment has been repeated many times, and it is clear that most proteins are capable of refolding correctly in dilute media in the absence of either other macromolecules or an added energy source. The same conclusion applies to the assembly of large multisubunit structures such as viruses and ribosomes. These observations were codified in the statement that proteins are capable of spontaneous self-assembly, the term “assembly” is used here to describe both the folding of individual polypeptide chains and their association into oligomers. The principle of protein self-assembly states that all the information for a protein chain to reach its correct conformation is encoded in its aminoacyl sequence and is an important corollorary of the central dogma of molecular biology. If self-assembly did not occur, molecular biology would be in deep trouble because it would have to be postulated that protein folding and assembly requires direction by an external agent. Generally, it was assumed that proteins also fold and assemble spontaneously inside the cell, but observations made in the late 1970s and early 1980s challenged this assumption. Some proteins were found to bind to preexisting native proteins before folding or after folding but before assembling. Such binding prevents the proteins from aggregating into nonfunctional structures, which is a problem aggravated by the macromolecular crowding found in most intracellular compartments (2, 3). The term “molecular chaperone” (often abbreviated to “chaperone”) was proposed to describe these helper proteins (4, 5), and the concept advanced that all cells require a chaperone function to prevent some proteins from misbehaving (6). Thus the original view that proteins fold and assemble inside the cell by a process of spontaneous self-assembly has been replaced by the concept that many proteins use an assisted self-assembly process. Note that the concept of protein self-assembly itself is unaffected by this change of paradigm—proteins contain all the information required for their correct folding and assembly. Their problem is to avoid aggregation.
Biologic Background
Protein aggregation is defined as the association of two or more polypeptide chains to form nonfunctional structures. Some aggregates are cytotoxic and cause neurodegenerative disease. Protein aggregation is at least a second-order process, so it is very sensitive to the concentration of interacting chains. Aggregation is also a highly specific process, so that only identical or very similar chains aggregate with one another. Because the free energy difference between folded and unfolded states is small, most folded proteins are stable only marginally and are easily unfolded into aggregation-prone states. All cells require a chaperone function to combat aggregation because two universal features of the intracellular environment, where proteins fold and assemble, aggravate both of these properties of aggregation. These features are the synthesis of proteins on polyribosomes and macromolecular crowding.
Polyribosomes
The linear deoxyribonucleotide sequence information encoded in genes is copied into a linear sequence of ribonucleotides in messenger RNA (mRNA) by the process of transcription. Ribosomes bind one at a time near to the 5' end of each mRNA molecule and translate the ribonucleotide base sequence into an aminoacyl sequence according to the genetic code. Each mRNA is bound to more than one ribosome at a time, forming a polyribosome, often abbreviated to polysome. Typically the rate of polypeptide chain synthesis is between 5 and 20 amino acid residues added per second, but the rate of protein folding is much faster. Thus, incomplete chains may fold into nonfunctional conformations before the entire chain is made. The fact that polypeptide chains are made by polysomes ensures that partly folded identical chains are within touching distance when are synthesized, which results in the possibility that chains may aggregate with one another. The advantage of simultaneously making several polypeptide chains from one molecule of mRNA has been earned at the risk of both misfolding and aggregation. Some chaperones reduce this risk by binding to incomplete and newly synthesized polypeptide chains before they complete their folding.
Macromolecular crowding
This term is used to highlight that the total concentration of macromolecules inside cells is very high, which is in the range 80-400 g/L. Such high total concentrations occur in all intracellular compartments where proteins fold and assemble. Generally, it is not appreciated that such a degree of crowding stimulates macromolecular association reactions, by levels that can be two to three orders-of-magnitude greater than those in uncrowded media (2, 3). Such association reactions include protein aggregation, and during some, but not all, protein refolding experiments, a fraction of the chains aggregate with one another, even though such experiments are almost always done in uncrowded media. Refolding experiments show the concentration and temperature-dependence of protein aggregation, so protein chemists solve this problem by reducing the concentration of chains and lowering the temperature. Cells cannot do this but employ molecular chaperones to combat the enhanced tendency to aggregate, which is caused by macromolecular crowding.
Origin of the molecular chaperone concept
The term “molecular chaperone” first appeared in 1978 to describe the properties of a nuclear protein called nucleoplasmin found in amphibian eggs (4). This protein assists the assembly of nucleosomes from histones and DNA that occurs when these eggs divide rapidly after fertilization. Nucleosomes are held together by the electrostatic bonds that are disrupted by high concentrations of salt. If the isolated histones and DNA are then incubated together at physiologic concentrations of salt, a dramatic failure of self-assembly exists—an insoluble aggregate forms instead of nucleosomes. Eggs solve this problem by producing large amounts of the acidic protein nucleoplasmin that binds to the histones, thereby reducing their high surface positive charge density and allowing their intrinsic self-assembly properties to predominate over their tendency to aggregate (4). The term “molecular chaperone” appears in the discussion of Reference 4 to describe the role of nucleoplasmin to prevent incorrect ionic interactions between histones and DNA but without forming part of the assembled nucleosome. The parallels with the properties of the human chaperone are obvious. Thus, the common perception that chaperones are concerned solely with assisting protein folding is incorrect and has been incorrect since this subject began. Recent work on the mechanism of action of those chaperones that assist assembly rather than folding has been reviewed (7).
In the 1980s, two reports suggested that some newly synthesized polypeptide chains bind to other preexisting proteins before they completed their assembly into functional complexes (8, 9). The first report led to a general concept of the molecular chaperone function, which was proposed as a useful and an accurate way to describe the function of a diverse class of proteins postulated to assist folding and assembly processes in all cells (5). The second report established that at least one family of chaperone functions at the level of protein folding rather than at the level of protein assembly (9). The term “molecular chaperone” is now in general use, but it is often applied uncritically by those unfamiliar with the origins of the concept or the definitions that are available.
Definitions
The molecular chaperone function currently is defined as the ability of all cells to prevent and/or reverse incorrect interactions that may result when potentially interactive surfaces are exposed to the environment (7). These surfaces occur on growing and newly released polypeptide chains, on mature proteins unfolded by environmental stresses, and on folded proteins in near-native and native conformations. The same concept applies to other macromolecules that can undergo incorrect interactions, especially RNA. Incorrect interactions are defined as those that result in products that do not carry out the functions for which they have been selected in evolution.
Molecular chaperones currently are defined as a large and diverse group of proteins that share the functional property of assisting the noncovalent folding/unfolding and the assembly/disassembly of other macromolecular structures but are not components of these structures when these are performing their normal biologic functions (7). This definition differs from the original one (6) in that includes unfolding and disassembly processes. This change was necessary because of the discovery that some chaperones are required for processes such as the remodeling of chromatin during fertilization, the disassembly of clathrin cages, and the dissolution of insoluble aggregates. Some molecular chaperones are also stress or heat shock proteins (hsp), because the requirement for chaperone function increases under conditions such as heat shock and other environmental stresses that cause some proteins to unfold and aggregate.
The above definition of molecular chaperone is entirely functional and contains no constraints on the mechanisms by which different chaperones may act. The term “noncovalent” is used to exclude those proteins that carry out posttranslational covalent modifications. Protein disulfide isomerise may seem to be an exception, but it is both a covalent modification enzyme and a molecular chaperone. It is helpful to think of a molecular chaperone as a function rather than as a molecule. Thus, no reason exists why a chaperone function should not be a property of the same molecule that has other functions. Other examples include peptidyl-prolyl isomerase, which possesses both enzymatic and chaperone activities in different regions of the molecule, and the alpha-crystallins, which combine two essential functions in the same molecule in the lens of the eye-contributing to the transparency and the refractive index required for vision as well as to the chaperone function that combats the loss of transparency as the protein chains aggregate with increasing age. The proteasome particle has a chaperone-like activity involved in unfolding proteins prior to their proteolysis. A recent proposal is that some chaperones also possess extracellular cell-cell signaling functions, based on many experiments that show that some exogenously applied chaperones elicit responses from cultured cells (10). It remains an open question as to how many other effects chaperones may have.
Classes of Chaperone
It is convenient to divide chaperones into two broad classes: those that assist the folding of newly synthesized or stress- denatured proteins and those that assist the assembly of oligomeric structures. Some of these chaperones are highly specific for their protein substrates, whereas others are not. Many proteins have been termed chaperones, usually on the basis of their in vitro properties, but no comprehensive list is available.
Chaperones involved in protein folding-small chaperones
Some chaperones assist the folding of both nascent chains bound to ribosomes, newly synthesised chains released from ribosomes (i.e., in both cotranslational and posttranslational modes), as well as mature proteins unfolded by environmental stresses and some membrane proteins (11-12-13). The chaperones working in these cotranslational and posttranslational modes are distinct, and they can be termed small and large chaperones, respectively, because this is a case where size is important for function. Small chaperones are less than 200 kDa in size and include trigger factor, nascent chain-asssociated complex, prefoldin, the hsp70 and hsp40 families, and their associated cochaperones. Cochaperones are defined as proteins that bind to chaperones to modulate their activity; they may or may not also be chaperones in their own right. Large chaperones are more than 800 kDa in size and include the thermosome in Archaea, GroE proteins in bacteria and the eukaryotic organelles evolutionarily derived from them, and the tailless complex polypeptide-1 (TCP-1) or TRiC complexes and associated cochaperones in the cytosol of Eukarya. The large chaperones are related evolutionarily and are referred to collectively as the chaperonins. In the endoplasmic reticulum lumen of eukaryotic cells, no large chaperones exist, but small chaperones do exist, such as BiP (a hsp70 homologue), calnexin, calreticulin, and protein disulphide isomerase that assist the folding of chains transported into the lumen after synthesis in the cytosol. Table 1 lists some of the chaperones that assist protein folding in various intracellular compartments, whereas Fig. 1 illustrates those chaperones that assist protein folding in the cytosol.
Small chaperones bind transiently to small hydrophobic regions (typically seven or eight residues long) on both nascent and completed newly synthesized chains, and thus, they prevent aggregation both during and after chain elongation by shielding these regions from one another (11, 12). Trigger factor (48 kDa) is the first chaperone to bind to nascent chains in prokaryotes because it is associated with the ribosomal large subunit at the tunnel from which the chains emerge (14). A cell of Escherichia coli contains about 20,000 copies of this chaperone, which is enough to bind to all nascent chains. Trigger factor shows peptidyl-prolyl isomerase activity and contains a hydrophobic groove that binds transiently to regions of the nascent chain enriched in aromatic residues. It binds to nascent chains as short as 57 residues and dissociates in an ATP-independent manner after the chain is released from the ribosome; this binding does not require prolyl residues in the nascent chain. The isomerase activity may provide a means to keep nascent chains that contain prolyl residues in a flexible state. The eukaryotic cytosol lacks the trigger factor, but its function may be replaced by that of a heterodimeric complex of 33-kDa and 22-kDa subunits, termed the nascent chain-associated complex. Like the trigger factor, this complex binds transiently to short nascent chains, but unlike the trigger factor, it does not possess peptidyl-prolyl isomerase activity.
Cells lacking the trigger factor show no phenotype, but this is because its function can be replaced by that of the other major small chaperone, hsp70 (15). The hsp70 family has many 70-kDa proteins distributed between the cytoplasm of bacteria and some Archaea, the cytosol of Eukarya, and eukaryotic organelles such as the endoplasmic reticulum, mitochondria, and chloroplasts. Some members are also stress proteins. Unlike the trigger factor, most hsp70 members do not bind to ribosomes but do bind to short regions of hydrophobic residues exposed on nascent and newly synthesized chains. Such regions occur statistically about every 40 residues and are recognized by a peptide-binding cleft in hsp70.
Most information is available about the hsp70 member in E. coli, termed DnaK. Like all hsp70 chaperones, DnaK contains an ATPase site and occupation of this site by ATP promotes rapid but reversible peptide binding. ATP hydrolysis tightens the binding through conformational changes in DnaK. The cycling of ATP between these states is regulated by a 41-kD cochaperone of the hsp40 family, termed DnaJ in E. coli, and GrpE, a nucleotide exchange factor that is a cochaperone but not a chaperone. DnaJ binds to DnaK through its J domain and increases the rate of ATP hydrolysis, which facilitates peptide binding. DnaJ, like all the hsp40 proteins, acts as a chaperone in its own right because it also binds to hydrophobic peptides. Thus, DnaK and DnaJ cooperate to bind each other to nascent chains; all hsp70 chaperones are thought to cooperate with hsp40 chaperones. The role of GrpE is to stimulate release of ADP from DnaK, which allows the latter to bind another molecule of ATP and to release the peptide. In the eukaryotic cytosol, the role of GrpE is fulfilled by an unrelated cochaperone called Bag-1. Some Archaea lack hsp70 proteins, but it is speculated that their role in protein folding may be replaced by that of an unrelated chaperone called prefoldin.
Enough DnaK is in a cell of E. coli for one molecule to bind to each nascent chain. DnaK binds to longer chains than the trigger factor, and so it probably binds after the trigger factor. When the gene for the trigger factor is deleted, the fraction of nascent and newly synthesized chains that bind to DnaK increases from about 15% to about 40%. However, removal of the genes for both the trigger factor and the DnaK in the same cell causes the aggregation of many newly synthesized chains and is lethal to the cell (15). This observation suggests that the redundancy of important control systems is as good a design principle for cells as it is for passenger planes. An exception to this principle is the GroEL chaperone (see the discussion in the next section), whose gene is essential for the survival of E. coli. Small chaperones function essentially by reducing the time that potentially interactive surfaces on neighboring chains are exposed by cycling these chains on and off until they have folded; they do not seem to change the conformation of the chains. The other major class of chaperones involved in protein folding, however, functions by a much more sophisticated mechanism enabled by its large size.
Table 1. Chaperones that assist protein folding
|
Family |
Other names Eukaryotes Prokaryotes |
Functions |
|
|
hsp100 |
hsp104, 78 |
ClpA/B/X |
Disassembly of oligomers and aggregates |
|
hsp90 |
hsp82, hsp83, Grp94 |
HtpG |
Regulation of assembly of steroid receptors and signal transduction proteins |
|
hsp70 |
hsc70, Ssa1-4, Ssb1-2, BiP, Grp75 |
DnaK, Hsc66 Absent from many Archaea |
Prevention of aggregation of unfolded protein chains |
|
Chaperonins |
hsp60, TRiC, CCT, TCP-1, Rubisco subunit binding protein |
GroEL, GroES |
Sequestering partly folded chains inside central cage to allow completion of folding in absence of other folding chains |
|
hsp40 |
Ydj1, Sis1, Sec63p, auxilin, zuotin, Hdj2 |
DnaJ |
Stimulation of ATPase activity of hsp70 |
|
Prefoldin |
GimC |
Absent from Bacteria Present in Archaea |
Prevention of aggregation of unfolded protein chains |
|
Trigger factor |
Absent from Eukarya |
Present |
Binding to nascent chains as they emerge from ribosome |
|
Calnexin, calreticulin |
Present |
Absent from prokaryotes |
Binding to partly folded glycoproteins; located in ER membrane and lumen, respectively |
|
Nascent chain-associated complex (NAC) |
Present |
Absent from prokaryotes |
Binding to nascent chains as they emerge from ribosome |
|
PapD Membrane chaperones |
Absent from Eukarya Shr3p Gsf2p Pho68p Chs7p |
Present in some |
Prevention of aggregation of subunits of pili Prevent aggregation of some integral polytopic membrane proteins |
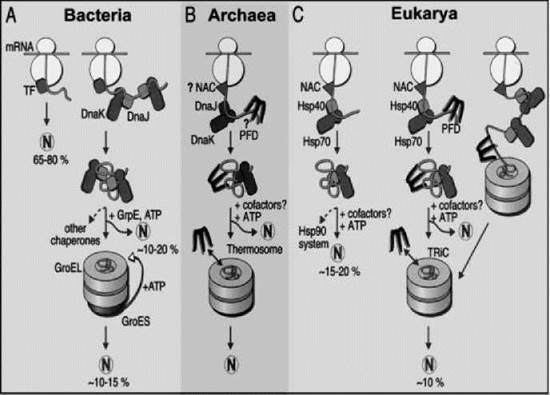
Figure 1. Models for the chaperone-assisted folding of newly synthesized polypeptides in the cytosol. (a) Bacteria. TF, trigger factor; N, native protein. Most nascent chains probably interact with TF, and most small proteins (about 65-80% of total chain types) may fold rapidly during synthesis without additional chaperone assistance. Longer chains (10-20% of total chain types) interact subsequently with DnaK and DnaJ and fold after one or several cycles of ATP-dependent binding and release. About 10-15% of total chains fold within the chaperonin GroEL/GroES system. GroEL does not bind to nascent chains and is likely to receive its substrates after their release from DnaK. (b) Archaea. PFD, prefoldin; NAC, nascent-chain associated complex. Only some archaeal species contain DnaK/DnaJ. The existence of a ribosome-bound NAC homologue and the binding of prefoldin to nascent chains have not been shown. (c) Eukarya. Like TF, NAC probably interacts with many nascent chains. Most smaller chains may fold without additional chaperone assistance. About 15-20% of chains reach their native states after assistance by hsp70 and hsp40, and a specific fraction of these are then transferred to hsp90. About 10% of chains are passed to the TRiC system in a reaction involving PFD. Reprinted from Reference (12) with permission from the American Association for the Advancement of Science.
Large chaperones involved in protein folding-the chaperonins
Most information is available for GroEL and GroES, the chaperonin and cochaperonin found in E. coli , but the general principles of their mechanism (Fig. 2) are suspected to apply also to the thermosome found in Archaea and to the TCP-1 complex (also called the TRiC or CCT complex) found in the eukaryotic cytosol. GroEL (800 kDa) consists of two heptameric rings of identical 57-kDa ATPase subunits stacked back to back, which contain a cage in each ring (17). The term “cage” is used because the walls surrounding each central cavity contain gaps, perhaps to allow water to enter and to exit. Each subunit contains three domains. The equatorial domain contains the nucleotide binding site and is connected by a flexible intermediate domain with the apical domain. The latter presents several hydrophobic side chains at the top of the ring oriented toward the cavity of the cage, which is an arrangement that permits either a partly folded polypeptide chain or a molecule of GroES to bind but prevents binding to another GroEL oligomer.
GroES is a single heptameric ring of 10-kDa subunits that cycle on and off either end of the GroEL in a manner regulated by the ATPase activity of GroEL. At any one time GroES is bound to only one end of GroEL, leaving the other end free to bind a partly folded polypeptide chain after its release from the ribosome. GroEL does not bind to nascent chains, but TCP-1 may do so. The two rings of GroEL are coupled by negative allostery so that only one ring at a time binds the nucleotide, but within each ring, the binding of the nucleotide is cooperative. When either ADP or ATP is bound to one GroEL ring, the GroES sits on top of this ring, which is now called the cis ring. The binding of GroES triggers a large rotation and upward movement of the apical domains, which results in an enlarged cage and a change in its internal surface properties from hydrophobic to hydrophilic. This enlarged cage can accommodate a single partly folded compact polypeptide chain up to about 60 kDa in size, perhaps depending on shape.
The reaction cycle starts with a GroEL-GroES complex containing ADP bound to the cis ring (Fig. 2, step 1). The hydrophobic residues on the apical domains on the other ring, now called trans, bind to hydrophobic residues exposed on a partly folded polypeptide chain, presumably after the release of small chaperones from this chain. GroES and ATP then bind to this ring, thereby converting it into a new cis ring and causing the release of GroES and ADP from the old cis ring (Fig. 2, step 2). This binding of GroES to the trans ring displaces the bound polypeptide into the cavity of the cage because some hydrophobic residues of the apical domains that bind the polypeptide are the same residues that bind GroES. The displaced chain lying free in the cavity of the cage now has 10-15 seconds to continue folding, a time that is set by the slow but cooperative ATPase activity of the seven subunits in each ring (Fig. 2, step 3). The chain continues its folding sheltered in a hydrophilic environment that contains no other folding chain. Many denatured polypeptide chains will fold completely within 15 seconds in the classic Anfinsen renaturing experiment carried out in a test tube instead of inside GroEL. It is for this reason that this mechanism has been called the “Anfinsen cage model.” The binding of ATP and GroES to the new trans ring triggers the release of GroES and ADP from the cis ring that contains the polypeptide chain, which allows the latter to diffuse out of the cage into the cytoplasm. If this chain has internalized its hydrophobic residues, it remains free in the cytoplasm (Fig. 2, step 4). But any chain that still exposes hydrophobic residues rebinds back to the same ring for another round of encapsulation (Fig. 2, step 5). Rebinding to the same ring rather than to the ring of another GroEL oligomer is favored by the crowding effect created by the high concentration of macromolecules in the cytoplasm and reduces the risk that partly folded chains will meet one another in the cytoplasm in a potentially disastrous encounter.
This model was proposed to explain the results of many ingenious in vitro experiments, but recent genetic studies confirm the importance of the Anfinsen cage mechanism in intact cells. Mutants in which the mechanism is prevented by blockage of the entrance to one ring of each GroEL oligomer are viable, but the cells form colonies only 10% the size of the wild-type colonies (18). That these mutants are viable at all suggests that the ring whose entrance is not blocked is acting like the small chaperones, i.e., reducing aggregation by binding and releasing from the hydrophobic regions on newly synthesized chains.
An unexpected, added advantage of the Anfinsen cage mechanism is that for proteins in a certain size range, encapsulation in the cage increases the rate of folding compared with the rate observed in free solution under conditions where aggregation is not a problem. Thus, the rate of folding of bacterial rubisco (50 kDa) is increased fourfold by encapsulation, whereas that of rhodanese (33 kDa) is not affected; alterations of the cavity size have different effects, depending on the size of the encapsulated protein (19). This effect can be explained in terms of a type of macromolecular crowding called confinement, in which the promixity of the walls of the confining cage stabilizes compact conformations more than extended ones, and so it enhances the rate of interactions that lead to compaction of the folding chain. Current estimates suggest that the fraction of newly synthesized polypeptide chains that bind in vivo to either hsp70 proteins or the chaperonins is in the range 10-20%. Whether most newly synthesized chains bind to other, as yet undiscovered, chaperones or fold unassisted because their sequences have evolved to avoid aggregation is unknown. Nor is it understood what determines that only a few defined polypeptides bind to GroEL in the intact cell. Some mycoplasmas lack genes for GroEL and GroES, perhaps because the limited number of proteins they contain do not include ones prone to aggregation.

Figure 2. Mechanism of action of the GroEL/GroES system in E.coli. P, unfolded polypeptide chain. N, native folded chain. For details, see text. Reprinted from Reference (16), Copyright 2003 with permission from Elsevier.
Chaperones assisting oligomeric assembly
The nuclear compartment contains high concentrations of negatively charged nucleic acids bound to positively charged proteins that together form chromatin and various ribonucleoprotein particles. Besides nucleoplasmin, several other chaperones assist the assembly and disassembly of these structures by preventing incorrect ionic interactions between them, whereas cytosolic assembly chaperones assist the assembly of ribosomes, pro- teasomes, and proteins such as hemoglobin (7, 20, 21). The continuing emphasis on the role of chaperones in protein folding may explain the relative dearth of information about assembly chaperones.
Common misconceptions
As with any new field, misconceptions abound. The literature contains statements of the form “chaperones fold proteins,” which suggests to the uninitiated that chaperones possess steric information essential for protein folding; such grammatical forms should be avoided. Molecular chaperones and stress proteins are sometimes used as though they were interchangeable categories, but although many proteins share both chaperone and stress functions, this is not the case. A common error is to use the term “chaperonin” as synonymous with the term “chaperone,” but it should be noted that the chaperonins are just one particular family of chaperone defined by sequence similarity, i.e., the family that contains GroEL, hsp60, and TCP-1. The occasional use of the nonsense term “molecular chaperonin” in some respectable journals suggests that some people use these terms casually and without reference either to their meaning or to their history. It should be obvious that the word “molecular” is used to qualify “chaperone” because in common usage, “chaperone” refers to a person. So the term “molecular chaperonin” is as nonsensical as the term “molecular immunoglobulin.”
Another common misconception is that molecular chaperones are necessarily promiscuous, i.e., that each assists the assembly of many different types of polypeptide chains. This assumption is true for the hsp70, hsp40, and GroE chaperonin families but not for hsp90, PapD, hsp47, Lim, Syc, ExbB, PrtM/PrsA, and prosequences, which are specific for their substrates. Similarly, it is not a universal property of chaperones that they hydrolyze ATP; hsp100, hsp90, and hsp70, and the chaperonins hydrolyse ATP, but they trigger factor, hsp40, prefoldin, calnexin, protein disulfide isomerise, and papD do not.
The term “chemical chaperone” has been proposed to describe small molecules such as glycerol, dimethylsulfoxide, and trimethylamine N-oxide that act as protein-stabilizing agents. This terminology is unfortunate and confuses students, because proteins are also chemicals. This term should be replaced by the term “kosmotrope” that physical chemists use to describe small molecules that stabilise proteins.
References
1. Anfinsen CB. Principles that govern the folding of protein chains. Science 1973; 181:223-230.
2. Ellis RJ. Macromolecular crowding; obvious but underappreciated. Trends Biochem. Sci. 2001; 26:597-603.
3. Ellis, RJ, Minton AP. Protein aggregation in crowded environments. Biol. Chem. 2006; 387:485-497.
4. Laskey RA, Honda BM, Finch JT. Nucleosomes are assembled by an acidic protein that binds histones and transfer them to DNA. Nature 1978; 275:416-420.
5. Ellis RJ. Proteins as molecular chaperones. Nature 1987; 328:378- 379.
6. Ellis RJ, Hemmingsen SM. Molecular chaperones: proteins essential for the biogenesis of some macromolecular structures. Trends Biochem. Sci 1989; 14:339-342.
7. Ellis RJ. Molecular chaperones: assisting assembly in addition to folding. Trends Biochem. Sci. 2006; 31:395-401.
8. Barraclough BR, Ellis RJ. Protein synthesis in chloroplasts. IX. Assembly of newly synthesized large subunits of ribulose bisphosphate carboxylase in isolated intact chloroplasts. Biochim. Biophys. Acta 1980; 608:19-31.
9. Ostermann J, Horwich AL, Neupert W, Hartl F-U. Protein folding in mitochondria requires complex formation with hsp60 and ATP hydrolysis. Nature 1989; 341:125-130.
10. Henderson B, Shamaei-Tousi A. Molecular chaperones: the unorthodox view. In: Henderson B, Graham A, Pockley, eds. Molecular Chaperones and Cell Signalling 2005. Cambridge University Press, New York.
11. Young JC, Agashe VR, Siegers K, Hartl FU. Pathways of chaperone-mediated protein folding in the cytosol. Nature Revs. Cell Biol. 2004; 5:781-790.
12. Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 2002; 295:1852-1858.
13. Kota J, Ljungdahl PO. Specialized membrane-localized chaperones prevent aggregation of polytopic proteins in the ER. J. Cell Biol. 2005; 168:79-88.
14. Kaiser CM, Chang H-C, Agashe VR, Laksmipathy SK, Etcherlls SA, Hayer-Hartl M, Hartl FU, Barral JM. Real-time observation of trigger factor function on translating ribosomes. Nature 2006; 444:455-460.
15. Deuerling E, Schulze-Specking A, Tomoyasu A, Mogk A, Bukau B. Trigger factor and DnaK cooperate in folding of newly synthesized proteins. Nature 1999; 400:693-696.
16. Ellis RJ. Protein folding: the importance of the Anfinsen cage. Curr. Biol. 2003; 13:R881-883.
17. Xu Z, Horwich AL, Sigler P. The crystal structure of the asymmetric GroEL-GroES-(ADP)7 chaperonin complex. Nature 1997; 388:741-750.
18. Farr GW, Fenton WA, Rospert S, Horwich AR. Folding with and without encapsulation by cis and trans-only GroEL-GroES complexes. EMBO J 2001; 22:3220-3230.
19. Tang Y-C, Chang H-C, Roeben A, Wischnewski D. Wischnewski N. Kerner MJ, Hartl FU, Hayer-Hart M. Structural features of the GroEL-GroES nanocage required for rapid folding of encapsulated protein. Cell 2006; 125:903-914.
20. Philpott A, Krude T, Laskey RA. Nuclear chaperones. Semin. Cell Dev. Biol. 2000; 11:7-14.
21. Akey CW, Luger K. Histone chaperones and nucleosome assembly. Curr. Opin. Struct. Biol. 2003; 13:6-14.
Further Reading
Csermely P, Vigh L, eds. Molecular aspects of the stress response; chaperones, membranes and networks 2007. Advances in experimental medicine and biology. Landes Bioscience and Springer Science + Business Media, LLC, New York.
Pearl LH, Prodromou C. Structure and mechanism of the hsp90 molecular chaperone machinery. Annu. Rev. Biochem. 2006; 75:271-294.
Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu. Rev. Biochem. 2005; 74:739-789.
See Also
Protein Folding, Chemical Biology of Diseases
Protein Misfolding
Proteins, Chemistry and Chemical Reactivity of
Proteins, Structure, Function and Stability
Self-Organization and Self-Assembly in Biology