CHEMICAL BIOLOGY
Attenuation, Control of Gene Expression by
Paul Babitzke, Department of Biochemistry and Molecular Biology, The Pennsylvania State University, University Park, Pennsylvania
Paul Gollnick, Department of Biological Science, State University at Buffalo, Buffalo, New York
doi: 10.1002/9780470048672.wecb030
Bacteria use a variety of strategies to regulate transcription elongation in response to changes in their environment. Several of these regulatory events are classified as transcription attenuation mechanisms, which involve regulated transcription termination to control expression of downstream genes. In most cases, two alternative RNA secondary structures, an antiterminator and an intrinsic terminator, share nucleotides in common such that their formation is mutually exclusive. Thus, the level of expression of the downstream genes depends on the frequency that the antiterminator forms in the nascent transcript. Transcription attenuation mechanisms in general are separated into two categories. The first category involves situations in which the action of a regulatory molecule promotes transcription termination, with the default situation being transcription readthrough. The second category involves situations in which the action of the regulatory molecule promotes transcription readthrough (antitermination), with the default situation being transcription termination. Transcription attenuation controls the expression of several amino acid biosynthetic and catabolic operons. These mechanisms are often controlled by the efficiency of leader peptide synthesis in Gram-negative bacteria, whereas Gram-positive organisms often use RNA binding proteins or tRNA molecules to determine whether the RNA will fold into the antiterminator or terminator structures. Several examples of transcription attenuation also exist in which an antisense RNA is responsible for controlling plasmid copy number. In addition, small metabolites participate in transcription attenuation mechanisms of a wide variety of bacterial genes by binding directly to the nascent transcript.
The ability of organisms to regulate gene expression in response to environmental signals is vital for their survival. Virtually every step involved in the synthesis, function, and degradation of macromolecules is a potential target for one or more regulatory events (reviewed in References 1-3). Several regulatory events that occur after transcription initiates are categorized as transcription attenuation mechanisms in which the extent of transcription termination is modulated to control the expression of downstream genes. Transcription attenuation allows the organism to modulate the extent of transcription readthrough past the terminator in response to changing environmental signals, thereby regulating expression of the downstream genes. Transcription attenuation mechanisms in general are divided into two categories. In one case, the default situation is transcription readthrough; the action of a regulatory (effector) molecule promotes termination. The default situation for the second category is transcription termination. In this case, the action of an effector molecule promotes transcription readthrough (antitermination) (1, 2). In each case, the genetic information required for transcription attenuation is encoded within a leader region located between the transcription start site and the first structural gene of the operon or between two genes within an operon (reviewed in Reference 4). This article is organized around the class of effector molecule that is used to sense environmental changes. These regulatory factors can be translating ribosomes, proteins, RNA molecules, or metabolites (Fig. 1). Examples are known in which each class of effector molecule promotes transcription termination or antitermination. One example of each of these general regulatory mechanisms is described in detail, whereas additional examples are included to illustrate particularly important points. A common theme that will emerge throughout this article is that synchronization of factor binding and/or RNA folding with RNA polymerase (RNAP) position is critical in all transcription attenuation mechanisms.
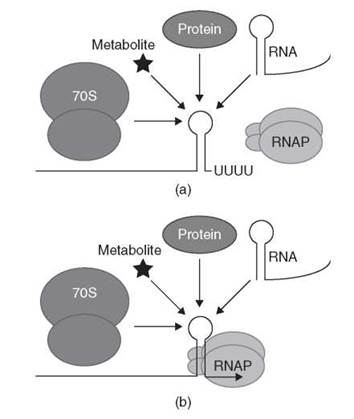
Figure 1. Generalized model for sensing regulatory effectors by nascent mRNA leader transcripts. Transcription attenuation mechanisms have been identified in which the nascent transcript interacts with a translating 70S ribosome, a protein, an RNA molecule or a small metabolite. (a) Binding of the effector molecule promotes transcription termination. (b) Binding of the effector molecule promotes transcription readthrough (antitermination). See text for details.
Transcription Attenuation Mediated by Ribosomes
The discovery of transcription attenuation was the first demonstration that organisms can exploit an RNA structure to modulate gene expression. The first examples of transcription attenuation were discovered for the Escherichia coli trp (tryptophan biosynthetic) and Salmonella typhimurium his (histidine biosynthetic) operons, although many other amino acid biosynthetic operons in Gram-negative bacteria are regulated in a similar fashion. Together, these discoveries established an important paradigm for regulation of gene expression after transcription initiates. It is now apparent that translating ribosomes can serve as the effector molecule for both mechanistic categories of transcription attenuation.
Transcription Termination Mediated by a Translating Ribosome
TrpR, which is a DNA binding repressor protein, regulates transcription initiation of the E. coli trpEDCBA operon. Under tryptophan limiting conditions, TrpR represses transcription initiation, whereas repression is relieved in the presence of excess tryptophan. Once transcription initiates the elongating transcription complex is subject to control by transcription attenuation (reviewed in References 5 and 6). The leader transcript can form three RNA secondary structures that are referred to as the pause hairpin, the antiterminator structure, and an intrinsic terminator hairpin. Because the antiterminator shares nucleotides in common with the terminator, their formation is mutually exclusive. The pause hairpin has two additional roles in this transcription attenuation mechanism; it serves as an anti-antiterminator structure that prevents antiterminator formation, and it codes for a leader peptide. A model of the E. coli trp operon transcription attenuation mechanism is presented in Fig. 2a.
Soon after transcription of the trp operon is initiated, the pause hairpin forms in the nascent transcript, which signals RNAP to pause. The paused RNAP complex allows sufficient time for a ribosome to initiate translation of a short open reading frame that encodes a 14-amino-acid leader peptide. The translating ribosome disrupts the paused RNAP complex such that transcription resumes, which thereby couples transcription and translation. At this point two different outcomes can occur depending on the level of tryptophan in the cell. Under tryptophan-limiting conditions, the aminoacylation of tRNATrp is inefficient such that the level of tryptophanyl-tRNATrp (also called charged-tRNATrp) is low. As a consequence, the translating ribosome stalls at either of two tandem Trp codons within the leader peptide coding sequence. Ribosome stalling at either Trp codon uncouples transcription from translation. As transcription proceeds, the antiterminator structure forms, which prevents formation of the overlapping terminator hairpin, resulting in transcription readthrough into the trp operon structural genes (Fig. 2a). Under tryptophan excess conditions, the concentration of tryptophanyl-tRNATrp is sufficient to allow efficient translation of the Trp codons within the leader peptide coding sequence. As a consequence, the ribosome continues translation to the end of the leader peptide unabated and dissociates from the mRNA. In this situation, the anti-antiterminator forms and prevents formation of the overlapping antiterminator. Because formation of the antiterminator is prevented, the overlapping terminator hairpin can form, which results in termination of transcription before RNAP reaches the trp operon structural genes. Thus, expression of the trp operon is decreased when the cell has an adequate supply of tryptophan.
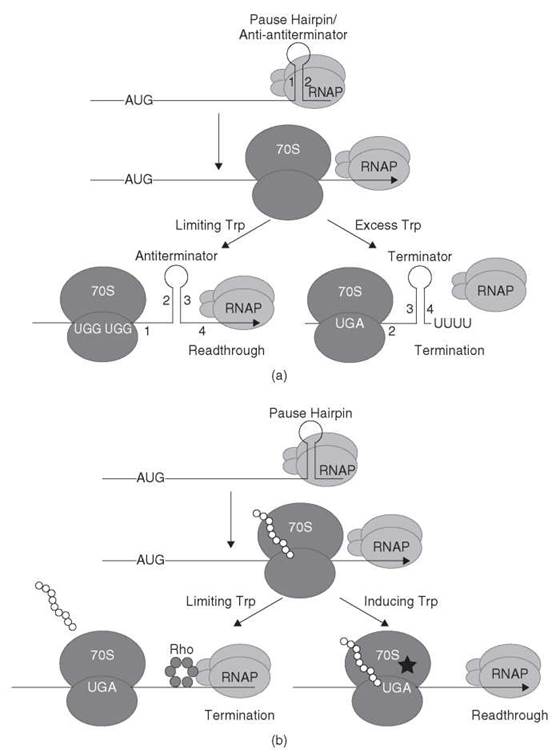
Figure 2. (a) Transcription attenuation model of the E. coli trpEDCBA operon. RNAP pauses after formation of the pause hairpin (stem-loop 1:2). RNAP pausing provides time for a ribosome to initiate translation of the leader peptide. In tryptophan-limiting conditions, the ribosome stalls at one of the tandem Trp codons, which thereby allows formation of the antiterminator structure (stem-loop 2:3), which results in transcription readthrough. In conditions of tryptophan excess, the ribosome reaches the leader peptide stop codon and dissociates. In this instance, formation of the anti-antiterminator blocks formation of the antiterminator, which thereby allows formation of the terminator hairpin (stem-loop 3:4), which causes transcription to terminate upstream from the trp operon structural genes. See text for details. [Adapted from the Encyclopedia of Molecular Biology (1999) with permission (1).] (b) Antitermination model of E. coli tnaCAB operon. RNAP pauses after formation of the pause hairpin. RNAP pausing provides time for a ribosome to initiate translation of the leader peptide. Under noninducing conditions (free tryptophan is not in excess), dissociation of the 70S ribosome at the tnaC stop codon exposes a rut site allowing Rho binding. Rho translocates to paused RNAP leading to transcription termination. Under inducing conditions (excess tryptophan), the combination of tryptophan binding and nascent peptide interaction with the ribosome exit channel causes the ribosome to stall at the tnaC stop codon because the nascent peptide cannot be cleaved from the peptidyl-tRNA. The stalled ribosome prevents Rho from binding to the nascent transcript that leads to transcription readthrough. Tryptophan is represented by a star. See text for details. [Adapted from the Encyclopedia of Molecular Biology (1999) with permission (2).]
Transcription Antitermination Mediated by a Translating Ribosome
E. coli and several other microorganisms have the capacity to degrade tryptophan to serve as a carbon and nitrogen source. The degradative tryptophanase operon (tnaCAB) of E. coli is regulated by catabolite repression and by an antitermination mechanism (reviewed in Reference 4). This antitermination mechanism involves translation of a 24-amino-acid leader peptide (tnaC) containing a crucial Trp codon (7, 8), RNAP pause site(s) between tnaC and tnaA (9), and Rho termination factor (10). A model that illustrates the tna antitermination mechanism is shown in Fig. 2b.
Under non-catabolite-repressing conditions, RNAP initiates transcription and continues until it reaches a pause site downstream of tnaC. Paused RNAP provides time for a ribosome to initiate translation of the leader peptide. Under tryptophan-limiting conditions, the translating ribosome reaches the UGA stop codon and is released, thereby exposing a rut (Rho utilization) site that immediately follows the stop codon. Rho binds to the rut site in the nascent tna transcript and translocates along the RNA in a 5' to 3' direction until it encounters paused RNAP, which ultimately leads to transcription termination upstream of tnaA. When cells are growing with inducing levels of tryptophan, the nascent TnaC peptide and a free molecule of tryptophan interact with the translating ribosome, which inhibits ribosomal peptidyl transferase activity such that peptidyl-tRNA remains bound to the ribosome (11-13). The stalled ribosome masks the rut site, which thereby prevents Rho interaction with the transcript. Eventually RNAP resumes transcription such that the structural genes encoding tryptophanase (tnaA) and a tryptophan permease (tnaB) are expressed (Fig. 2b).
The mechanism of ribosome stalling during translation of tnaC has been investigated in considerable detail. The nascent leader peptide (TnaC) interacts with ribosomal protein L22 and 23 S rRNA in the narrow region of the ribosome exit channel (14). Ribosome recognition of the TnaC peptide results in specific binding of free tryptophan in the ribosome, which inhibits cleavage and the subsequent release of the nascent leader peptide (15, 16).
Transcription Attenuation Mediated by Proteins
Although the transcription attenuation mechanism of the E. coli trp operon served as a paradigm for several years, it is now apparent that several classes of effector molecules can be responsible for sensing and responding to environmental signals. Numerous examples exist in which RNA binding proteins perform this important task. In some instances, the presence of a specific metabolite leads to activation of the RNA binding protein such that it can bind to its RNA targets. In other examples, the metabolite leads to inactivation of the RNA binding protein. Despite this fundamental difference, several features of the corresponding mechanisms are remarkably similar to those described above. One clear distinction that is worthy of particular mention is that the mechanisms that rely on RNA binding proteins do not require coupling of transcription and translation. As a consequence, this general regulatory strategy has been identified in several eukaryotic organisms in which transcription and translation occur in separate cellular compartments (reviewed in Reference 3).
Transcription Termination Mediated by an RNA Binding Protein
The transcription attenuation mechanism for the trpEDCFBA operon in the Gram-positive organism Bacillus subtilis differs dramatically from that described for E. coli. Most notably, translation of a leader peptide is not involved in the B. subtilis mechanism. Instead, an RNA binding protein called TRAP (trp RNA-binding attenuation protein) is responsible for sensing the level of tryptophan in the cell and for the decision of whether to terminate transcription in the 5' leader region, or to readthrough into the trp operon structural genes (reviewed in References 17 and 18). Transcription initiation of the trp operon seems to be constitutive, which suggests that the > 1000-fold regulation in response to tryptophan occurs after transcription initiates. A transcription attenuation model for the B. subtilis trp operon is depicted in Fig. 3a.
The B. subtilis trp leader transcript contains inverted repeats that can form mutually exclusive antiterminator and intrinsic terminator structures. Soon after transcription initiates, RNAP can pause at the nucleotide just preceding the critical overlap between theses two alternative structures, presumably to provide additional time for TRAP binding (19, 20). Under tryptophan excess conditions, tryptophan-activated TRAP binds to 11 closely spaced (G/U)AG repeats in the nascent transcript. As transcription proceeds, the RNA wraps around the protein ring with each triplet repeat interacting with several amino acids on adjacent subunits in the protein (21). Bound TRAP prevents formation of the antiterminator, which allows formation of the overlapping terminator structure. Thus, transcription halts in the leader region before RNAP reaches the trp operon structural genes. An RNA hairpin called the 5' stem-loop (5'SL) also interacts with TRAP; however, the mechanism of TRAP binding to this structure is distinct from TRAP interaction with the (G/U)AG repeats (22-24). It seems that TRAP-5'SL RNA interaction increases the rate in which TRAP binds to the nascent trp leader transcript, which thereby increases the efficiency of termination (24). Under tryptophan-limiting conditions, TRAP is not activated and does not bind to the nascent trp operon transcript. As a consequence, formation of the antiterminator prevents formation of the terminator, which results in transcription readthrough into the trp operon structural genes (Fig. 3a).
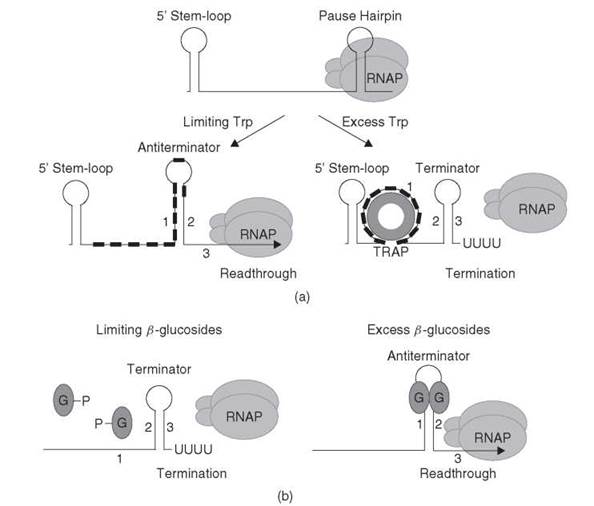
Figure 3. (a) Transcription attenuation model for the B. subtilis trpEDCFBA operon. RNAP pauses after formation of the pause hairpin. RNAP pausing may provide additional time for tryptophan-activated TRAP to bind to the nascent transcript. In tryptophan-limiting conditions, TRAP does not bind to the 5'SL or the (G/U)AG repeats (small black rectangles). Thus, formation of the antiterminator (stem-loop 1:2) results in transcription readthrough. In conditions of tryptophan excess, tryptophan-activated TRAP binds to the 5'SL and the triplet repeats. Bound TRAP blocks or disrupts formation of the antiterminator structure, which thereby allows formation of the terminator hairpin (stem-loop 2:3), which causes transcription to terminate upstream from the trp operon structural genes. See text for details. (b) Antitermination model for the E. coli bglGFB operon. Under noninducing conditions (limiting β-glucosides), the phosphorylated BglG monomers cannot bind to the nascent transcript. Thus, formation of a terminator hairpin (stem-loop 2:3) causes transcription to terminate. In the presence of p-glucosides, the BglG monomers are dephosphorylated, which allows them to dimerize. BglG dimers bind and stabilize the antiterminator structure (stem-loop 1:2), which prevents formation of the overlapping terminator, and transcription continues into the structural genes. Note that overlapping antiterminator and terminator hairpins are found in the leader region and between the bglG and the bglF coding sequences. See text for details.
The crystal structure of B. subtilis TRAP in complex with tryptophan shows that this protein consists of 11 identical subunits arranged in a ring (25). The TRAP oligomer is composed of 11 seven-stranded antiparallel β-sheets, with each β-sheet containing 4 β-strands from one subunit and 3 β-strands from the adjacent subunit. L-tryptophan binds between each adjacent subunit with the indole ring buried in a hydrophobic pocket. Nine hydrogen bonds are formed among the amino group, the carboxyl group, and the indole nitrogen of each bound tryptophan, with amino acids residing in two loops on adjacent subunits. The hydrogen bonds formed among Thr30, Gln47, and Thr49 and tryptophan are essential for TRAP function. Conversely, the hydrogen bond between Ser53 and the carboxyl group of tryptophan is dispensable for TRAP activity (26).
The mechanism of TRAP activation by tryptophan is not known because the structure of nonliganded TRAP (apo-TRAP) has not been determined, although it is known that tryptophan binding is not required for oligomerization of the protein (27). NMR studies of TRAP in both the nonliganded and the activated states demonstrate that apo-TRAP is more flexible than the tryptophan-activated protein (28), which imply that reduced flexibility plays an important role in altering the RNA binding activity of TRAP.
Four amino acid residues within each TRAP subunit (E36, K37, K56, and K58) are critical for RNA binding (29). Three of these residues form KKR motifs that are aligned on the perimeter of the B. subtilis and the homologous B. stearother-mophilus TRAP ring. Several crystal structures of B. stearother-mophilus TRAP-RNA complexes show that the RNA wraps around the TRAP protein ring with the bases pointing toward the protein, such that the majority of the hydrogen bond interactions are with the RNA bases and there are no contacts to phosphates (21, 30). The only direct hydrogen bond to the RNA backbone is to the 2' OH of the ribose of the third G in each repeat. K37 forms a hydrogen bond with the A of each GAG repeat, whereas K56, R58, and E36 hydrogen bond with the G in the third position of each repeat. The side chain of K37 also forms hydrophobic interactions with the base in the first residue of each repeat. The spacer bases do not make specific contacts with TRAP, which is consistent with the lack of sequence conservation in spacers of natural TRAP binding sites and with findings that indicate that their composition in general is not critical for TRAP binding.
A model of TRAP-trp leader RNA interaction that encompasses several recent findings is as follows. Once a sufficient number of triplet repeats emerge from RNAP (e.g., three or four repeats), TRAP binds to the nascent trp leader transcript by contacting the 5'SL and the emerging triplet repeats. The nature of these contacts orients the protein so the RNA easily can wrap around TRAP’s periphery as transcription continues. TRAP sequesters the remaining repeats in a 5' to 3' directionality as they emerge from RNAP (31, 32). As the tenth and eleventh repeats bind, the 5'SL is displaced as a consequence of the geometry of the TRAP-RNA complex in which TRAP completely is encircled by RNA (24). In conjunction with RNAP pausing, this RNA binding mechanism ensures that TRAP binds in time to block formation of the antiterminator structure.
Transcription Antitermination Mediated by an RNA Binding Protein
RNA binding proteins are responsible for mediating antitermination of several catabolic operons in bacteria. BglG, SacT, SacY, and LicT belong to a family of more than 50 proteins that regulate gene expression by a common antitermination mechanism. In this mechanism, the bound protein prevents formation of an intrinsic transcription terminator just upstream from the regulated gene(s) (reviewed in Reference 33). A general model for the bgl antitermination mechanism in E. coli is shown in Fig. 3b.
The E. coli bglGFB operon encodes all functions necessary for the regulated uptake and utilization of aromatic β-glucosides. Because transcription from the bgl promoter is constitutive, it seems that BglG, which is a dimeric RNA binding protein, is solely responsible for controlling expression of this operon in response to P-glucosides (33). In the absence of β-glucosides, BglG does not bind RNA and most transcripts terminate at one of two intrinsic terminators present in the leader region upstream of bglG and in the intercistronic region between bglG and bglF. At inducing levels of β-glucosides, BglG binds to one of two antiterminators in the nascent bgl transcript that partially overlap their respective terminators. Binding of BglG prevents formation of the terminators by stabilizing the antiterminators. Thus, expression of bglG only requires transcription readthrough past the first terminator, whereas expression of bglF and bglB requires readthrough past both terminators.
The RNA binding activity of BglG is regulated by BglF, which is a membrane protein that is responsible for both transport of β-glucosides and reversible phosphorylation of BglG. In the absence of β-glucosides, BglF phosphorylates a histidine residue in BglG (H208), which prevents BglG from dimerizing and binding to the nascent bgl transcript. In the presence of β-glucosides, BglF dephosphorylates BglG, which allows dimerization and RNA binding. Phosphorylation of both β-glucosides and BglG is accomplished by transfer of the phosphate group from Cys24 in BglF (34). Thus, under conditions in which β-glucoside levels are high, the phosphate group can be transferred from BglG back to Cys24 in BglF, and subsequently to β-glucosides. It seems that BglF recruits BglG to the cell membrane in the absence of β-glucosides such that the system can respond rapidly to the presence of the stimulating sugar (35).
Expression of two sucrose utilization operons in B. subtilis, sacPA and sacB, is induced by sucrose by similar transcription antitermination mechanisms; antitermination is mediated by the RNA binding proteins SacT and SacY, respectively (33). The structure of the RNA binding domain of SacY has been solved by both NMR (36) and X-ray crystallography (37). The domain exists as a dimer with each monomer consisting of a four-stranded antiparallel β-sheet. The structure of the RNA-binding domain from the LicT antiterminator protein in complex with a 29 base RNA shows that LicT binds mostly through hydrophobic and stacking interactions with the RNA and that binding “clamps” the RNA so as to stabilize the antiterminator structure (38).
Transcription Attenuation Mediated by RNA
RNA is a third class of effector molecule that can direct the decision to terminate transcription or to allow transcription to continue into the coding sequence of genes. In some systems, binding of antisense RNA molecules to the nascent leader transcript promotes transcription termination. These antisense RNAs typically act in cis as they are transcribed from an overlapping gene in the opposite direction. In addition, tRNA molecules are known to interact specifically with nascent leader transcripts of a large number of genes, which thereby promote transcription readthrough. As is the situation with RNA binding proteins, coupling of transcription and translation is not a component of these regulatory mechanisms.
Transcription Termination Mediated by an RNA Molecule
Antisense RNA-mediated transcription attenuation controls the copy number of several Gram-positive plasmids (reviewed in References 39 and 40). In the case of the Streptococcus agalactiae plasmid, pIP501, the antisense RNA (RNA III) inhibits expression of repR, which is the gene encoding the essential RepR initiator protein, by binding to the nascent repR leader transcript (RNA II) (Fig. 4a). This interaction promotes formation of an intrinsic terminator upstream of the repR coding sequence. In the absence of RNA III-RNA II interaction, an alternative structure forms in the nascent RNA II transcript that blocks formation of the terminator. Although this alternative structure is more complex, it is functionally analogous to the antiterminator structures described above.
Interaction of RNA III with RNA II is initiated by the formation of a so-called kissing complex between single-stranded loops of both molecules, followed by propagation of the intermolecular RNA helix (41). The kissing interaction involves base pairing between loops III and IV from RNA III with loops II and I of RNA II, respectively (42). Moreover, loop L1 of RNA II contains a YUNR (Y = C or U; N = A, C, G or U; R = A or G) “U-turn” motif that is important for the kissing interaction; mutations that disrupt the U-turn motif result in higher plasmid copy number (43). Although RNA III and RNA II share extensive sequence complementarity, complete pairing is not necessary to promote transcription termination of RNA II. The finding that inhibition of transcription readthrough is considerably faster than stable complex formation suggests that inhibition precedes stable pairing (41). Furthermore, complete pairing only occurs at a low frequency because pairing of complementary folded RNAs often arrests at the stage of stable binding intermediates (41).
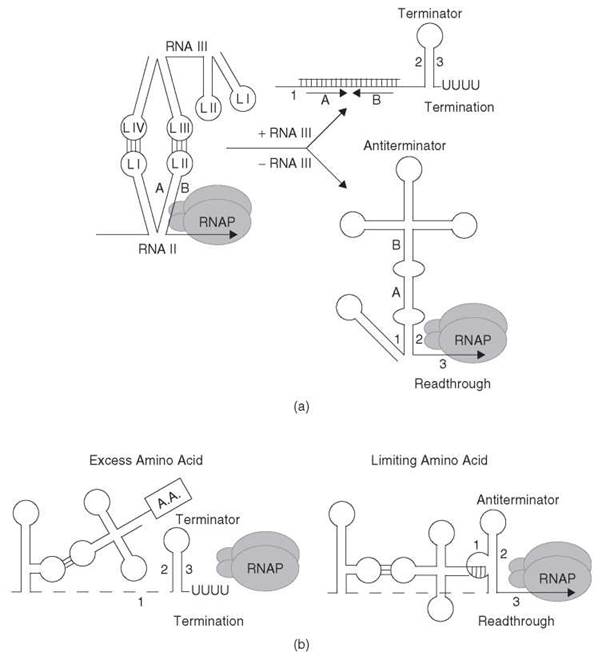
Figure 4. (a) Transcription attenuation model of S. agalactiae plasmid pIP501-encoded repR. When the concentration of RNA III is low, the absence of RNA III interaction allows formation of the antiterminator structure (stem-loop 1:2), which blocks formation of the terminator that leads to transcription readthrough. When RNA III is abundant, the kissing loop interaction between loops L III and L IV of RNA III with loops L I and L II within the nascent RNA II (repR) transcript blocks formation of the antiterminator, which promotes formation of the terminator hairpin (stem-loop 2:3) and transcription termination in the repR leader region. The positions of the residues that correspond to L II of RNA II (stem-loop A:B) are included in each structure for clarity. See text for details. [Adapted from the Encyclopedia of Molecular Biology (1999) with permission (1).] (b) General T-box model of tRNA-mediated antitermination. Under excess amino acid conditions, the amino acid on charged tRNA does not allow interaction of the tRNA with the discriminator sequence in the nascent transcript. As a consequence, formation of the terminator (stem-loop 2:3) halts transcription in the leader region. Under limiting amino acid conditions, uncharged tRNA binds to the nascent transcript via anticodon-specifier sequence base pairing as well as by base pairing between the 3' end of tRNA and the discriminator region. This second interaction stabilizes the antiterminator structure (stem-loop 1:2), which results in transcription readthrough. The tRNA is shown as a cloverleaf structure, and a boxed A.A. attached to the tRNA indicates it is aminoacylated. See text for details. (Adapted from Reference 45 with permission.)
Transcription Antitermination Mediated by an RNA Molecule
In several bacterial species, uncharged tRNA serves as the effector molecule in controlling expression of several aminoacyl-tRNA synthetase genes and a few amino acid biosynthetic operons by a common mechanism termed T-box antitermination. Most of these genes are from Gram-positive bacteria, although a few examples have been identified in Gram-negative organisms as well (reviewed in References 44 and 45). Expression of these genes is induced specifically by starvation for the cognate amino acid. A generalized model of the T-box antitermination mechanism is shown in Fig. 4b.
The untranslated leader region of each operon contains several conserved structural elements preceding an intrinsic transcription terminator. When the amino acid pool in the cell is sufficiently high, transcription terminates prematurely in the leader region upstream from the coding sequences. In addition to the conserved secondary structures, a critical 14-nt sequence called the T box is present in each leader region. Another structure of the leader region, which forms when the T-box base pairs with the 5' side of the terminator stem, functions as an antiterminator, which allows transcription to read through into the structural genes (46).
Another critical feature of the leader region of T-box-controlled genes is a trinucleotide sequence that corresponds to a codon for the appropriate amino acid involved in regulating each operon. For example, in tyrS, which encodes tyrosyl-tRNA synthetase, the leader contains a UAC tyrosine codon (46). This codon-like sequence is always present within an internal loop of a conserved RNA structure. This triplet is designated the “specifier sequence” because it specifically interacts with the anticodon of the cognate tRNA. A second base pairing interaction between the acceptor end of uncharged tRNA and the complementary sequence in the T box leads to stabilization of the antiterminator structure. The antiterminator side bulges contain a UGGN sequence, where N corresponds to a variable T-box position that covaries with the residue preceding the CCA acceptor end of the cognate tRNA (47, 48). The presence of the amino acid on charged tRNA prevents this second interaction from taking place (49). Although tRNA interaction with the specifier and UGGN sequences are firmly established, substantial genetic data suggest that additional tRNA-mRNA contacts take place (44).
Reconstitution of the B. subtilis glyQS T-box mechanism in vitro with purified components suggests that additional protein factors are not required for tRNA-mediated antitermination (50). Furthermore, the finding that charged tRNA can compete with uncharged tRNA for mRNA binding indicates that the ratio of uncharged:charged tRNA is monitored by the nascent transcript, rather than simply the concentration of uncharged tRNA (49). Although codon-anticodon interaction takes place at least 100 nt upstream from the terminator, nascent transcripts that extend just upstream of the terminator are still competent for tRNA binding and antitermination (49). Thus, it is possible that RNAP pausing participates in this regulatory mechanism by providing additional time for tRNA binding, although a regulatory pausing event has not been firmly established (51).
Under starvation conditions for the corresponding amino acid, the ratio of uncharged-to-charged tRNA is relatively high. In this case, efficient interaction of uncharged tRNA with both the specifier and the discriminator sequence elements of the nascent leader transcript promotes formation of the antiterminator structure, which allows transcription to proceed into the coding region. The resulting increase in the level of tRNA synthetase presumably allows more efficient charging of the scarce amino acid. As the concentration of the amino acid increases, an increasing proportion of nascent transcripts will interact with charged tRNA. In these instances, formation of the terminator hairpin will cause RNAP to terminate transcription in the leader region.
Transcription Attenuation Mediated by Metabolites
The most recently identified class of transcription attenuation mechanism involves direct sensing of the effector molecule by the nascent transcript (52-54). These RNA sensors control metabolically diverse pathways. As for the other attenuation and antitermination mechanisms discussed thus far, recognition of the particular effector molecule occurs with the appropriate affinity and high specificity required for precise control of gene expression.
Transcription Termination Mediated by a Small Metabolite
In B. subtilis, 26 genes grouped into 11 operons constitute the S-box regulon. These genes are involved in sulfur metabolism as well as in the biosynthesis of methionine, cysteine, and S-adenosylmethionine (SAM) (55-57). SAM, the effector of the S-box regulon, binds to the nascent transcript with exquisite specificity; the closely related SAM analog S-adenosylhomocysteine binds with much lower affinity to the RNA targets and does not promote transcription termination in vitro (54, 58, 59). A simplified model of SAM-mediated attenuation is presented in Fig. 5a. Under limiting SAM concentrations, the nascent S-box transcript adopts an antiterminator structure that allows transcription to proceed into the structural gene(s). However, under conditions of SAM excess, SAM binds to the nascent transcript and stabilizes an anti-antiterminator structure, which blocks formation of the antiterminator. As a consequence, the terminator hairpin forms and transcription halts in the leader region.
High-resolution structural information has been obtained for several RNA sensors (56, 60-63). A common feature of the leader RNAs is that they form tertiary structures in which single-stranded loops base pair with one another (pseudoknot). For example, the tertiary structure of the SAM-sensing RNA contains a pseudoknot that occurs between the loop of an RNA hairpin and the single-stranded junction between two other helices (64). This pseudoknot seems to stabilize the global architecture of the structure and probably facilitates SAM recognition (56). Another common characteristic of these structures is engulfment of the ligand by the binding pocket of the folded RNA (63).
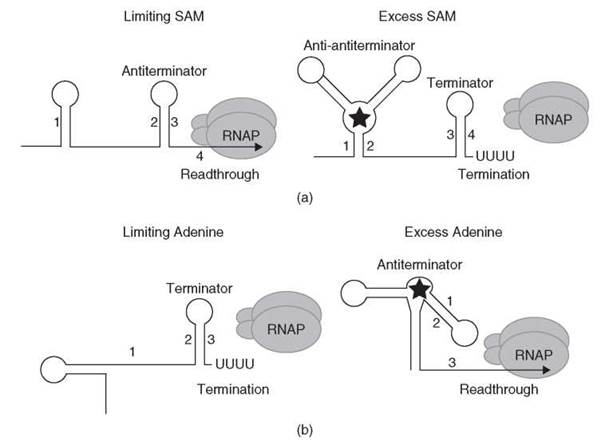
Figure 5. (a) Simplified SAM-mediated transcription attenuation model. Under limiting SAM conditions, the antiterminator forms (stem-loop 2:3) and transcription continues into the structural gene(s). Under conditions where SAM is in excess (depicted as a star), SAM binds to the nascent transcript and stabilizes an anti-antiterminator structure (stem-loop 1:2), which blocks formation of the antiterminator. As a consequence, the terminator hairpin forms (stem-loop 3:4) and transcription halts in the leader region. See text for details. (b) Antitermination model of the B. subtilis pbuE gene. Under limiting adenine conditions, the terminator forms (stem-loop 2:3) and transcription halts in the leader region. Under conditions where adenine is in excess (depicted as a star), adenine binds to the nascent transcript and stabilizes the antiterminator structure, which includes stem-loop 1:2. Formation of the antiterminator prevents formation of the terminator and transcription proceeds into the coding sequence. See text for details.
Transcription Antitermination Mediated by a Small Metabolite
Adenine sensing by the nascent B. subtilis pbuE (ydhL) transcript constitutes the most recently described antitermination mechanism (63-65). Although this is the only reported example of this type of antitermination mechanism, it is likely that additional examples will be identified. A model that depicts this antitermination mechanism is shown in Fig. 5b. Under limiting adenine conditions, an intrinsic terminator hairpin forms in the nascent pbuE transcript, which causes RNAP to terminate transcription in the leader region. In the presence of excess adenine, adenine binds to the nascent transcript and stabilizes formation of an alternative antiterminator structure, which thereby allows transcription to continue into the pbuE structural gene (65). Adenine-mediated activation of pbuE expression makes biologic sense as it encodes an apparent purine efflux pump (66). Deletions of the adenine-binding domain of the leader RNA caused constitutive expression of pbuE and conferred resistance to 2-fluoroadenine, which suggests that PbuE can pump this adenine analog out of the cell (65, 66). Analog binding studies demonstrated that the binding affinity of adenine and 2-aminopurine are similar, whereas the affinity of 2,6-diaminopurine is considerably higher. However, the association rate was faster for adenine compared with the two purine analogs. This finding supports a model in which regulation depends on the kinetics of ligand binding and the rate of transcription, rather than simple binding affinity (67).
The structure of the pbuE leader RNA is similar to the guanine-dependent RNA sensor in the xpt leader transcript (65). It was shown that a single C to U substitution in the loop of a triple helical junction swapped the xpt aptamer specificity from guanine to adenine. Importantly, the pbuE leader contains a U in the identical position. These results led to the hypothesis that this U residue in the pbuE leader base-paired with adenine, whereas the C residue in the xpt transcript paired with guanine This hypothesis was verified by NMR structural studies. Moreover, it was determined that adenine binding to pbuE leader RNA involved a base triple with two U residues, which includes the previously proposed uridine (68).
Concluding Remarks
The discovery of transcription attenuation over 30 years ago led to the realization that mRNAs serve a purpose beyond simply functioning as a conduit of information from DNA to protein. Indeed, the discovery of transcription attenuation established for the first time that RNA structure can modulate gene expression. It is now abundantly clear that expression of many genes is controlled by several different mechanisms after transcription initiates. Furthermore, it is apparent that transcription attenuation occurs by a variety of mechanisms that differ in the nature of the effector molecule (translating ribosome, RNA binding protein, RNA molecule, small metabolite), whether the effector promotes transcription termination (attenuation) or transcription readthrough (antitermination), as well as the structure of the RNA target in the nascent transcript. Although each mechanism described in this review contains key differences, it is also apparent that all but one of these mechanisms share an important feature: the presence of mutually exclusive antiterminator and terminator structures. One point of view is that these RNA-based regulatory mechanisms are relics of an RNA world in which both the storage of genetic information and the metabolic function were carried out by RNA molecules. Conversely, it is conceivable that these regulatory mechanisms have evolved more recently. In either case, it is apparent that what was once viewed as a biologic quirk is widespread and has been adapted to suit the physiologic needs of perhaps every organism.
Acknowledgments
The authors thank the members of their laboratories, both past and present, for stimulating discussion and discovery. This work was supported by grants GM052840 to P. B. and GM062750 to P.G. from the National Institutes of Health.
References
1. Babitzke P, Gollnick P. Attenuation of transcription. In: Encyclopedia of Molecular Biology. Creighton TE, ed. 1999. John Wiley & Sons, New York. pp. 216-220.
2. Gollnick P, Babitzke P. Antitermination control of gene expression. Encyclopedia of Molecular Biology. In: Creighton TE, ed. 1999. John Wiley & Sons, New York. pp. 173-177.
3. Gollnick P, Babitzke P. Transcription attenuation. Biochim. Biophys. Acta. 2002; 1577:240-250.
4. Henkin TM, Yanofsky C. Regulation by transcription attenuation in bacteria: how RNA provides instructions for transcription ter- mination/antitermination decisions. Bioessays. 2002; 24:700-707.
5. Yanofsky C. Attenuation in the control of expression of bacterial operons. Nature. 1981; 289:751-758.
6. Yanofsky C. Transcription attenuation. J. Biol. Chem. 1988; 263:609-612.
7. Gollnick P, Yanofsky C. tRNA(Trp) translation of leader peptide codon 12 and other factors that regulate expression of the tryptophanase operon. J. Bacteriol. 1990; 172:3100-3107.
8. Gish K, Yanofsky C. Evidence suggesting cis action by the TnaC leader peptide in regulating transcription attenuation in the tryptophanase operon of Escherichia coli. J. Bacteriol. 1995; 177:7245-7254.
9. Gong F, Yanofsky C. A transcriptional pause synchronizes translation with transcription in the tryptophanase operon leader region. J. Bacteriol. 2003; 185:6472-6476.
10. Stewart V, Landick R, Yanofsky C. Rho-dependent transcription termination in the tryptophanase operon leader region of Escherichia coli K-12. J. Bacteriol. 1986; 166:217-223.
11. Gong F, Ito K, Nakamura Y, Yanofsky C. The mechanism of tryptophan induction of tryptophanase operon expression: tryptophan inhibits release factor-mediated cleavage of TnaC-peptidyl- tRNA(Pro). Proc. Natl. Acad. Sci. U.S.A. 2001; 98:8997-9001.
12. Gong F, Yanofsky C. Instruction of translating ribosome by nascent peptide. Science. 2002; 297:1864-1867.
13. Cruz-Vera LR, Gong M, Yanofsky C. Changes produced by bound tryptophan in the ribosome peptidyl transferase center in response to TnaC, a nascent leader peptide. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:3598-3603.
14. Cruz-Vera LR, Rajagopal S, Squires C, Yanofsky C. Features of ribosome-peptidyl-tRNA interactions essential for tryptophan induction of tna operon expression. Mol. Cell. 2005; 19:333-343.
15. Cruz-Vera LR, New A, Squires C, Yanofsky C. Ribosomal features essential for tna operon induction: tryptophan binding at the peptidyl transferase center. J Bacteriol. 2007; 189:3140-3146.
16. Gong M, Cruz-Vera LR, Yanofsky C. Ribosome recycling factor and release factor 3 action promotes TnaC-peptidyl-tRNA dropoff and relieves ribosome stalling during tryptophan induction of tna operon expression in Escherichia coli. J. Bacteriol. 2007; 189:3147-3155.
17. Babitzke P. Regulation of transcription attenuation and translation initiation by allosteric control of an RNA-binding protein: the Bacillus subtilis TRAP protein. Curr. Opin. Microbiol. 2004; 7:132-139.
18. Gollnick P, Babitzke P, Antson A, Yanofsky C. Complexity in regulation of tryptophan biosynthesis in Bacillus subtilis. Annu. Rev. Genet. 2005; 39:47-68.
19. Yakhnin AV, Babitzke P. NusA-stimulated RNA polymerase pausing and termination participates in the Bacillus subtilis trp operon attenuation mechanism in vitro. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:11067-11072.
20. Yakhnin AV, Yakhnin H, Babitzke P. RNA polymerase pausing regulates translation initiation by providing additional time for TRAP-RNA interaction. Mol. Cell. 2006; 24:547-557.
21. Antson AA, Dodson EJ, Dodson G, Greaves RB, Chen X, Gollnick P. Structure of the trp RNA-binding attenuation protein, TRAP, bound to RNA. Nature 1999; 401:235-242.
22. Sudershana S, Du H, Mahalanabis M, Babitzke P. A 5' RNA stem-loop participates in the transcription attenuation mechanism that controls expression of the Bacillus subtilis trpEDCFBA operon. J. Bacteriol. 1999; 181:5742-5750.
23. Du H, Yakhnin AV, Dharmaraj S, Babitzke P. trp RNA-binding attenuation protein-5' stem-loop RNA interaction is required for proper transcription attenuation control of the Bacillus subtilis trpEDCFBA operon. J. Bacteriol. 2000; 182:1819-1827.
24. McGraw A, Bevilacqua PC, Babitzke P. TRAP-5' stem-loop interaction increases the efficiency of transcription termination in the Bacillus subtilis trpEDCFBA operon leader region. RNA. In press.
25. Antson AA, Otridge J, Brzozowski AM, Dodson EJ, Dodson GG, Wilson KS, Smith TM, Yang M, Kurecki T, Gollnick P. The structure of trp RNA-binding attenuation protein. Nature 1995; 374:693-700.
26. Yakhnin AV, Trimble JJ, Chiaro CR, Babitzke P. Effects of mutations in the L-tryptophan binding pocket of the Trp RNA-binding attenuation protein of Bacillus subtilis. J. Biol. Chem. 2000; 275:4519-4524.
27. Li PT, Scott DJ, Gollnick P. Creating hetero-11-mers composed of wild-type and mutant subunits to study RNA binding to TRAP. J. Biol. Chem. 2002; 277:11838-11844.
28. McElroy C, Manfredo A, Wendt A, Gollnick P, Foster M. TROSY-NMR studies of the 91kDa TRAP protein reveal allosteric control of a gene regulatory protein by ligand-altered flexibility. J. Mol. Biol. 2002; 323:463-473.
29. Yang M, Chen X, Militello K, Hoffman R, Fernandez B, Baumann C, Gollnick P. Alanine-scanning mutagenesis of Bacillus subtilis trp RNA-binding attenuation protein (TRAP) reveals residues involved in tryptophan binding and RNA binding. J. Mol. Biol. 1997; 270:696-710.
30. Hopcroft NH, Manfredo A, Wendt AL, Brzozowski AM, Gollnick P, Antson AA. The interaction of RNA with TRAP: the role of triplet repeats and separating spacer nucleotides. J. Mol. Biol. 2004; 338:43-53.
31. Barbolina MV, Li X, Gollnick P. Bacillus subtilis TRAP binds to its RNA target by a 5' to 3' directional mechanism. J. Mol. Biol. 2005; 345:667-679.
32. Barbolina MV, Kristoforov R, Manfredo A, Chen Y, Gollnick P. The rate of TRAP binding to RNA is crucial for transcription attenuation control of the B. subtilis trp operon. J. Mol. Biol. 2007; 370:925-938.
33. Amster-Choder O. The bgl sensory system: a transmembrane signaling pathway controlling transcriptional antitermination. Curr. Opin. Microbiol. 2005; 8:127-134.
34. Chen Q, Arents JC, Bader R, Postma PW, Amster-Choder O. BglF, the sensor of the E. coli bgl system, uses the same site to phosphorylate both a sugar and a regulatory protein. EMBO J. 1997; 16:4617-4627.
35. Lopian L, Nussbaum-Shochat A, O’Day-Kerstein K, Wright A, Amster-Choder O. The BglF sensor recruits the BglG transcription regulator to the membrane and releases it on stimulation. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:7099-7104.
36. Manival X, Yang Y, Strub MP, Kochoyan M, Steinmetz M, Aymerich S. From genetic to structural characterization of a new class of RNA-binding domain within the SacY/BglG family of antiterminator proteins. EMBO J. 1997; 16:5019-5029.
37. van Tilbeurgh H, Manival X, Aymerich S, Lhoste JM, Dumas C, Kochoyan M. Crystal structure of a new RNA-binding domain from the antiterminator protein SacY of Bacillus subtilis. EMBO J. 1997; 16:5030-5036.
38. Yang Y, Declerck N, Manival X, Aymerich S, Kochoyan M. Solution structure of the LicT-RNA antitermination complex: CAT clamping RAT. EMBO J. 2002; 21:1987-1997.
39. Brantl S. Antisense-RNA regulation and RNA interference. Biochim. Biophys. Acta. 2002; 1575:15-25.
40. Brantl S. Regulatory mechanisms employed by cis-encoded antisense RNAs. Curr. Opin. Microbiol. 2007; 10:102-109.
41. Brantl S, Wagner EG. Antisense RNA-mediated transcriptional attenuation occurs faster than stable antisense/target RNA pairing: an in vitro study of plasmid pIP501. EMBO J. 1994; 13:3599- 3607.
42. Heidrich N, Brantl S. Antisense RNA-mediated transcriptional attenuation in plasmid pIP501: the simultaneous interaction between two complementary loop pairs is required for efficient inhibition by the antisense RNA. Microbiology 2007; 153:420-427.
43. Heidrich N, Brantl S. Antisense-RNA mediated transcriptional attenuation: importance of a U-turn loop structure in the target RNA of plasmid pIP501 for efficient inhibition by the antisense RNA. J. Mol. Biol. 2003; 333:917-929.
44. Henkin TM. Control of transcription termination in prokaryotes. Ann. Rev. Genet. 1996; 30:35-57.
45. Grundy FJ, Henkin TM. Regulation of gene expression by effectors that bind to RNA. Curr. Opin. Microbiol. 2004; 7:126-131.
46. Grundy FJ, Henkin TM. tRNA as a positive regulator of transcription antitermination in B. subtilis. Cell. 1993; 74:475-482.
47. Grundy FJ, Rollins SM, Henkin TM. Interaction between the acceptor end of tRNA and the T box stimulates antitermination in the Bacillus subtilis tyrS gene: a new role for the discriminator base. J. Bacteriol. 1994; 176:4518-4526.
48. Yousef MR, Grundy FJ, Henkin TM. Structural transitions induced by the interaction between tRNA(Gly) and the Bacillus subtilis glyQS T box leader RNA. J. Mol. Biol. 2005; 349:273-287.
49. Grundy FJ, Yousef MR, Henkin TM. Monitoring uncharged tRNA during transcription of the Bacillus subtilis glyQS gene. J. Mol. Biol. 2005; 346:73-81.
50. Grundy FJ, Winkler WC, Henkin TM. tRNA-mediated transcription antitermination in vitro: codon-anticodon pairing independent of the ribosome. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:11121-11126.
51. Grundy F, Henkin T. Kinetic analysis of tRNA-directed transcription antitermination of the Bacillus subtilis glyQS gene in vitro. J. Bacteriol. 2004; 186:5392-5399.
52. Mironov AS, Gusarov I, Rafikov R, Lopez LE, Shatalin K, Kreneva RA, Perumov DA, Nudler E. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell 2002; 111:747-756.
53. Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 2002; 419:952-956.
54. McDaniel BA, Grundy FJ, Artsimovitch I, Henkin TM. Transcription termination control of the S box system: direct measurement of S-adenosylmethionine by the leader RNA. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:3083-3088.
55. Nudler E, Mironov AS. The riboswitch control of bacterial metabolism. Trends Biochem. Sci. 2004; 29:11-17.
56. Sashital DG, Butcher SE. Flipping off the riboswitch: RNA structures that control gene expression. ACS Chem. Biol. 2006; 1:341-345.
57. Grundy FJ, Henkin TM. The S box regulon: a new global transcription termination control system for methionine and cysteine biosynthesis genes in gram-positive bacteria. Mol. Microbiol. 1998; 30:737-749.
58. Epshtein V, Mironov AS, Nudler E. The riboswitch-mediated control of sulfur metabolism in bacteria. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:5052-5056.
59. Winkler WC, Nahvi A, Sudarsan N, Barrick JE, Breaker RR. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat. Struct. Biol. 2003; 10:701-707.
60. Montange RK, Batey RT. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature 2006; 441:1172- 1175.
61. Thore S, Leibundgut M, Ban N. Structure of the eukaryotic thiamine pyrophosphate riboswitch with its regulatory ligand. Science 2006; 312:1208-1211.
62. Serganov A, Polonskaia A, Phan AT, Breaker RR, Patel DJ. Structural basis for gene regulation by a thiamine pyrophosphatesensing riboswitch. Nature 2006; 441:1167-1171.
63. Tucker BJ, Breaker RR. Riboswitches as versatile gene control elements. Curr. Opin. Struct. Biol. 2005; 15:342-348.
64. McDaniel BA, Grundy FJ, Henkin TM. A tertiary structural element in S box leader RNAs is required for S-adenosylmethionine- directed transcription termination. Mol. Microbiol. 2005; 57:1008-1021.
65. Mandal M, Breaker RR. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat. Struct. Mol. Biol. 2004; 11:29-35.
66. Johansen LE, Nygaard P, Lassen C, Agerso Y, Saxild HH. Definition of a second Bacillus subtilis pur regulon comprising the pur and xpt-pbuX operons plus pbuG, nupG (yxjA), and pbuE (ydhL). J. Bacteriol. 2003; 185:5200-5209.
67. Wickiser JK, Cheah MT, Breaker RR, Crothers DM. The kinetics of ligand binding by an adenine-sensing riboswitch. Biochemistry. 2005; 44:13404-13414.
68. Noeske J, Richter C, Grundl MA, Nasiri HR, Schwalbe H, Wohnert J. An intermolecular base triple as the basis of ligand specificity and affinity in the guanine- and adenine-sensing riboswitch RNAs. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:1372-1377.
Further Reading
Weisberg RA, Gottesman ME. Processive antitermination. J. Bacteriol. 1999; 181:359-367.
This excellent review article summarizes the mechanisms responsible for processive antitermination in lambdoid bacteriophages, which includes the classic N-mediated antitermination of lambda and factor-independent antitermination of the related HK022 phage. This work was not included in this article because of space considerations.
See Also
mRNA Untranslated Regions (UTRs)
Protein-Nucleic Acid Interactions
Small Molecule-Nucleic Acid Interactions
Transcription, Activators and Repressors of
Transcription, Initiation of