CHEMICAL BIOLOGY
Glycan Sequencing and Data Integration for Glycomics
Rahul Raman and Ram Sasisekharan, Department of Biological Engineering, Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, Massachusetts
doi: 10.1002/9780470048672.wecb247
Databases and informatics tools are central to rapid acceleration in the progress of the genomics and proteomics fields. Glycomics, an emerging paradigm in the age of postgenomics, involves understanding the structure-function relationship of glycans in fundamental biological processes. In comparison with genomics and proteomics, the advancement of glycomics has faced unique challenges in the pursuit of developing tools, biological readouts, and an informatics platform to investigate glycan structure-function relationships. These challenges are because of the structural diversity and heterogeneity in glycans that develops from their complex nontemplate biosynthesis. This review focuses on areas of technologies for sequencing glycans and the importance of developing a bioinformatics platform to integrate the diverse data sets generated using the different technologies to allow a systems approach to glycan structure-function relationships. Although aspects of glycan analysis and integration of glycomics data sets have been covered in earlier reviews, this review brings these aspects together for both linear and branched glycans.
Glycosylation, or the attachment of complex carbohydrates (glycans) to proteins, is the most extensive and complex form of posttranslational modification and is required for the functional diversity to generate extensive phenotypes from a limited genotype. Glycans can be classified broadly as linear and branched, based on their chemical structure. Branched glycans (Fig. 1) are present as N-linked and O-linked glycosylation on glycoproteins or on glycolipids (1, 2). The majority of linear sugars are glycosaminoglycans (GAGs) that contain long polymers of sulfated disaccharide repeat units that are O -linked with a core protein to form a proteoglycan aggregate (3) (Fig. 1). The known intracellular roles of protein glycosylation include facilitating and stabilizing the protein folding process and targeting proteins to various intracellular compartments (1, 2). The glycans present on the glycoproteins and proteoglycans at the cell surface and the cell-extracellular matrix (ECM) interface are instead in an environment of many proteins including growth factors, cytokines, immune receptors, enzymes, and others. The ubiquitous distribution of glycans at the cell-ECM interface results in their involvement in the dynamic interplay between the cell and its environment. Glycans thus play a central role in fundamental biological processes such as development, cancer, infection, and immunity (4-8). The numerous biological roles of glycans are attributed to their specific interactions with a variety of proteins. Therefore, glycans modulate protein activity at the cell-ECM interface.
Informatics approaches are emerging as critical tools to accelerate understanding of the structure-function relationships of genes and proteins in fundamental biological processes. Such approaches involve storing, integrating, manipulating, analyzing, and mining sequence, structure, and biological-function information on genes and proteins at the molecular, cellular, tissue, and higher levels. The need for informatics tools continues to grow in the face of rapid data evolution from single-gene or protein level to complex high-throughput data sets on multiple genes or proteins at multiple levels (molecular, cellular, etc.). In the case of glycans, comprehending structure-function (or sequence-activity) relationships presents unique challenges when compared with genes and proteins. The biosynthesis of glycans is a non-template-driven process that involves the coordinated expression of several glycosyltransferases (and sulfotransferases in the case of GAGs), some of which have additional tissue-specific isoforms (1-3, 5). The complex biosynthesis and the lack of a proofreading machinery lead to an inherent heterogeneity and large diversity of glycan structures. Owing to their mode of biosynthesis, ubiquitous subcellular distribution, and the glycoprotein diversity that develops from one or more glycosylation sites, glycans additionally always need to be considered as a heterogeneous mixture of different chemical structures when isolated from cells and tissues. These challenges have hampered the development of analytical techniques that accurately can define the chemical structures on the ensemble of glycans isolated from glycoproteins, cells, or tissues. An understanding of the biochemical basis for glycan-protein interactions is complicated more by the multivalency and graded affinity that involve an ensemble of glycans making multiple contacts with multivalent protein binding sites (3, 9, 10).
In light of the above complexities, it is evident that developing an informatics platform is critical to decode the structure-function relationships of glycans. Informatics-based approaches are especially key to decode the sequence of glycans because no automated sequencing strategies are available similar to those that are found for DNA and proteins. Additionally, a need has developed to cut across diverse data, ranging from glycan biosynthesis, glycan characterization, and glycan-protein interactions, to take an integrated system or glycomics approach to delineate glycan structure-function relationships. This review is divided into two parts to provide a perspective on the development and application of informatics tools for glycans. The first part covers the critically needed informatics tools that sequence glycans to decode their sequence diversity. The second part covers the development of an informatics platform that bridges multiple data sets collected using different technologies to provide a systems framework for understanding glycan structure-function. These arguments are illustrated using examples of both linear and branched sugars.

Figure 1. Linear and branched glycans. Shown in figure are the different classes of glycan in their symbol nomenclature. The directionality of the glycans is from the nonreducing end at the top to the reducing end at the bottom with the arrows indicating the extension at the nonreducing side. The links between monosaccharides contain the anomeric configuration of the monosaccharide (α-alpha and β-beta] and the oxygen atom in the reducing end monosaccharide to which it is linked. The common terminal sugars in N-linked and O-linked branched glycans are shown in a dotted box (α3/6 indicates α3 or α6 link for the sialic acid). In the case of GAGs, representative chains of heparan sulfate (HS), chondroitin sulfate (CS), and dermatan sulfate (DS) are shown.
Decoding Glycan Diversity: Strategies for Sequencing Glycans
The complete set of glycans in a given cell (analogous to the genome or proteome of a cell) includes different types of gly- coconjugates such as glycolipids, glycoinositolphospholipids, glycans attached to proteins in the form of glycoproteins, pepti- doglycans, and proteoglycans. Decoding the complete chemical structure (analogous to the primary sequence of DNA and protein) of each of these glycans in the entire repertoire mentioned presents a formidable challenge. Many efforts have focused on the isolation and characterization of glycans from glycoproteins (for branched glycans) and proteoglycans (for linear GAGs).
Isolation of glycans from glycoproteins
Two distinct approaches characterize glycans on glycoproteins. The first approach involves releasing the glycans and characterizing the structures of the entire ensemble of these released glycans. The second approach instead fragments the entire glycoprotein to glycopeptides and characterizes the peptide and glycan component of each glycopeptide. The second approach involves characterization of glycoproteins in terms of the distinct glycan structures at each of their glycosylation sites. Glycans can be released from glycoproteins by using either chemical methods, such as hydrazinolysis (N-linked) and β-elimination (O-linked), or enzymatic methods that use PNGase F or A (N-linked). Currently, no generic enzymatic methods are available to release O-linked glycans. The enzyme O-glycanase, however, is frequently used in combination with sialidase to release simple O-linked oligosaccharides. The characterization of glycopeptides derived from the entire set of glycoproteins in a given cell is more difficult. The primary challenge lies with the relatively low abundance of glycosylated peptides when compared with nonglycosylated peptides. Methods such as lectin affinity chromatography (11) and 2D-SDS-PAGE separation (12) have helped to address the abundance issue and have led to the enrichment and homogenization of specific glycopeptides.
Mass spectrometry methods for glycan analysis
Mass spectrometry-(MS)-based approaches are used extensively to characterize glycans and glycopeptides. The overall strategy of MS-based approaches involves the ionization of the glycan sample to obtain the m/z (or mass to charge ratio) of the parent glycans. One or more sets of monosaccharide compositions, including several hexoses (Hex), N-acetylhexosamines (HexNAc), deoxyhexoses (dHex), N-acetyl neuraminic acids (Neu5Ac) and N-glycolyl neuraminic acids (Neu5Gc), would satisfy a given mass ion. For each set of compositions, one or more defined glycan topologies (such as biantennary, triantennary, etc.) without explicit assignment of the anomeric configuration and link between sugars is assigned. The application of rules from the known biosynthetic pathways of the glycans dramatically restricts this degeneracy. In some cases (particularly for lower mass values), a given mass peak could correspond uniquely to a single set of compositions and a defined glycan topology. Additional characterization to resolve the above degeneracy requires fragmentation of the parent mass ions (MS2) and the repetition of this process for subsequent fragment ions (MS3, MS4,..., MSn). The fragmentation can be directed in a specific fashion by employing different fragmentation techniques, ionization mode (positive or negative), or charge state of the parent ion and type of the MS instrumentation.
The most extensively used MS-based approaches for characterizing glycans are matrix assisted laser desorption-ionization (MALDI MS) and electrospray ionization (ESI MS) (13-15). The MALDI-MS approach uses a matrix to excite the sample and typically produces singly charged mass ions, which can be detected in positive or negative ion mode depending on the charge. The selection of the matrix and use of different derivatizations of the reducing end of the glycans improves ionization and increases sensitivity in both positive and negative ion mode. The ESI-MS approach is amenable to coupling with liquid chromatography-based separation of glycans where both positive and negative ion modes provide excellent analysis of mixtures containing both neutral and acidic glycans. Unlike MALDI-MS, ESI-MS generates multiple charged ions for the same glycan, which provides more informative fragmentation spectra. A highly sensitive approach using Fourier transform ion cyclotron mass spectrometry (FT-ICR-MS) has also been used to characterize different glycans including glycolipids (16).
Tools for interpretation of analytical data to sequence glycans
It is clear that the MS and fragmentation analysis of a large set of glycans isolated from glycoproteins or cells or tissues would result in complex data sets, which challenge the manual interpretation capability. The development of informatics tools for glycans is motivated by this need to analyze large data sets and has focused on facilitating the interpretation of data sets to determine the sequence of glycans (Table 1) (17-28). It should be noted that a trade-off occurs between the accurate characterization of the exact glycan structure and high-throughput analysis of several glycans in a given sample. In the case of a typical high-throughput data set such as MALDI-MS profile of glycans isolated from cells/tissues, a set of compositions and glycan topologies are assigned to a given mass peak based on biosynthetic constraints. The GlycoMod program (17) available on the ExPASy website provides an automatic calculation of possible monosaccharide compositions for a given parent ion mass. The Cartoonist software (18) provides an automated annotation of a MALDI-MS glycan profile with glycan topologies selected from a library of topologies generated using biosynthesis rules. These programs provide a quick read of the graphical representation of glycan topologies for an entire repertoire of glycans in a biological sample.
The next level of characterization involves matching the fragmentation patterns of the parent ion from the MS2 (and possibly higher order MS3,..., MSn) data to reference data sets and deducing the most likely glycan structures based on this data. The common reference data sets are those derived from the theoretical fragmentation of known glycan structures stored in different glycan structures databases. Glyco-Search-MS software is an online tool available to the public developed at the German Cancer Research Center (http://www.glycosciences.de) that matches MS2 data to theoretical fragments derived from a glycan structures database. The EuroCarb DB initiative has recently developed the Glyco-Peakfinder (20), which is a tool that rapidly annotates glycan MS spectra and MSnfragmentation spectra. An important feature of this tool is its ability to calculate the various fragment ions (singly and multiply charged) from glycosidic and cross-ring cleavages in a single cycle in parallel with the MS profile and thus to provide a fast and complete annotation of the whole spectrum. The GlycoWorkbench is another tool developed by EuroCarb DB initiative that provides the user- friendly graphical interface “GlycanBuilder” (21) to construct glycan structures and compute their theoretical fragmentation patterns. Unlike the standardized protein sequence databases (SwissProt and NCBI Protein) used in peptide fragmentation matching for proteomics, no such databases represent the best collection of all known glycans at this point of time. The possible glycan structures that match the fragmentation data could vary, therefore, depending on the glycan structures database used. The data are biased toward the known structures in the databases and thus present a challenge in the identification of any novel structures using these approaches. To circumvent some of these challenges, other approaches have been used to generate the reference data sets. The mass of the parent ion is used as a constraint to generate possible theoretical compositions. This set of compositions then is used to generate possible glycan structures (using biosynthesis rules in some cases) that have theoretical fragmentation pathways matched with the MSn data tree. Reference data sets have also been generated from experimental fragmentation profiles of well-characterized glycan structures or oligosaccharide fragments (29, 30). The STAT (23), StrOligo (24), and OSCAR (25) programs use these approaches. As the library of structures is derived theoretically from possible compositions for a given mass, the search space for matching the fragmentation pattern of a given parent ion is dramatically increased and leads to the higher probability of uncovering novel glycan structures. The limitations of these approaches include the need for extensive fragmentation data to resolve the numerous structural isomers developing from the theoretical structures and handling the computational intensity needed for parent ions with high molecular weights (>3000 Daltons).
The above MS methods and data interpretation tools are valuable to obtain rapidly the most probable structures of an entire ensemble of glycans isolated from cells and tissues. Determining the exact structure of each glycan (in a mixture), including the anomeric configuration of each sugar and the specific link ab initio, remains a formidable task. One approach toward this task employs exo-glycosidases that specifically cleave monosaccharides from the nonreducing end of a glycan. The HPLC chromatographic profile of parent glycan sample and the shifts in peaks that result from treatment with the exo-glycosidases are matched with reference chromatographic profiles of known glycan structures using software tools to derive the exact structure of glycans in the sample (26, 31). Nuclear magnetic resonance (NMR) spectroscopy is another valuable tool to obtain link information in glycans (32). The characteristic NMR chemical shifts and coupling constants of various glycans in literature have been compiled in databases to facilitate the assignment of glycan structures based on NMR data (Table 1). Such approaches for fine structure characterization of glycans have limitations when considering biological samples composed of a diverse mixture of glycans. The requirement of high sample amounts (in the case of NMR) and multiple steps of exo-glycosidase treatment for larger glycan mixtures complicate the use of these techniques for high-throughput analysis.
In the case of linear sugars such as GAGs, because of the heterogeneity in the chain length and sulfation pattern (Fig. 1), their isolation from biological samples results in a mixture of different chain lengths and compositions. To determine the sequence of individual GAG oligosaccharides in a mixture, it is necessary to subfractionate them based on size and charge and then to purify these fractions to homogeneity. The chain length of a homogeneous GAG oligosaccharide isolated in this manner is typically 4-8 disaccharide repeat units. It is difficult to determine the exact sequence of a GAG chain (particularly for highly sulfated GAGs) that is greater than 10 disaccharide repeat units. Given that GAG-protein binding specificity typically involves chain length of 2-5 disaccharide units, sequencing these chains does provide important information on the structure-function relationships of GAGs (3).
The two main areas for generating tools for the characterization of GAGs have been the development of chemical and enzymatic tools for the controlled and specific depolymerization or modification of a GAG chain and the development of analytical tools based on chromatography, electrophoresis, MS, and NMR techniques (see Reference 3 for details). Informatics-based methods have also been developed to capture the information density of GAGs and to enable the application of data from a combination of tools as constraints to sequence GAGs (27, 28). The underlying rationale of these approaches is to begin with a comprehensive master list of GAG sequences that represents the entire sequence space for a given chain length and overall composition (monosaccharide or disaccharide units). The data from a series of GAG depolymerization/modification profiles (mass spectrometry, electrophoresis, and chromatographic) serve as constraints to reduce this master list in an iterative fashion to the final sequence or set of probable sequences that satisfy all the constraints. These strategies provide an unbiased approach to sequencing GAGs through elimination of sequences from a master list that do not satisfy the experimental constraints.
Table 1. Resources for sequencing glycans
|
Analytical Tool/software |
Reference/online Access |
Overall logic |
|
GlycoMod |
www.expasy.org/tools/glycomod/ (17) |
Predicts glycan monosaccharide compositions (that satisfy biosynthesis rules) from mass spectrometric data. |
|
Cartoonist |
Goldberg et al. (18) |
Automated annotation of MALDI-MS using a library of glycan topologies derived from glycan biosynthesis rules. Applicable mainly to mammalian N-linked glycans. |
|
GlycoSearch-MS |
www.dkfz.de/spec/glycosciences.de/sweetdb/ms/ (19) |
Matches MS2 data to a theoretical fragmentation of structures in a glycan structure database. |
|
Glyco-Peakfinder |
www.dkfz.de/spec/EuroCarbDB/applications/ms-tools/ GlycoPeakfinder/GlycoPeakfinder.action (20) |
Automated annotation of MS and MSn fragmentation spectra with different types of fragment ions (glycosidic and cross ring cleavages). The composition of mass ion peaks can be used to search glycan structures database for possible structures. |
|
GlycanBuilder/GlycoWorkbench |
www.dkfz-heidelberg.de/spec/EUROCarbDB/GlycoWorkbench/ (21) |
An integrated platform to build glycan structures, simulate their fragmentation, and enable automated annotation of MS profile along with the MSn fragmentation spectra. |
|
GlycosidIQ |
www.glycosuite.com Joshi et al. (22) |
Matches MS2 data to a theoretical fragmentation of a database of reported glycan structures in literature. Not open access and requires subscription. |
|
Saccharide Topology Analysis Tool STAT |
Gaucher et al. (23) |
Reference data set containing set of all possible glycan topologies satisfying set of compositions generated based on parent ion mass. Mass of MSn fragmentation pattern matched to fragmentation of reference structures. |
|
StrOligo |
Ethier et al. (24) |
Compositional mass predicts possible structures based on known N-linked mammalian biosynthetic pathways, which are theoretically fragmented and matched to experimental data. |
|
Oligosaccharide |
glycome.unh.edu/tools/GlySpy/ Lapadula et al. (25) |
Theoretical MSn fragmentation trees are used to predict possible glycan topologies matching compositions for a given mass derived from a fragment composition finder database. The inferential rules to match MSnfragmentation are constructed de novo, so this tool does not match with glycan structures database and thus is better suited to identify novel structures. |
|
Subtree Constraint Algorithm OSCAR |
||
|
Automated interpretation of HPLC glycan profiles |
Rudd et al. (26) |
Parent glycan samples are treated with a combination of exoglycosidase specifically to remove selected monosaccharides from nonreducing end. Peak shifts are interpreted based on database of HPLC migration times for known glycan standards. |
|
CASPER |
www.casper.organ.su.se/casper/ |
Enables determination of glycan structure based on 1H or 13C NMR chemical shifts, component and link analysis. |
|
Glycosciences NMR DB |
www.glycosciences.de/sweetdb/nmr/ |
Database of 1H and 12C NMR shifts of glycans with tools to search glycans using NMR shifts or estimate shift for a given glycan. |
|
PEN-MALDI sequencing of GAGs
|
Venkataraman et al. (27) |
Disaccharide composition from capillary electrophoresis (CE) and MALDI-MS profiles of chemical and enzymatic degradation or modification of GAGs are applied as constraints using a computational framework to reduce the entire GAG sequence space for that given composition to converge on the final sequence or a set of sequences that satisfy all the constraints. |
|
PEN-NMR sequencing of GAGs |
Guerrini et al. (28) |
Similar to PEN-MALDI strategy but uses monosaccharide composition and link abundance from NMR along with disaccharide composition from CE as constraints to converge on the final sequence or set of sequences that satisfy all the constraints. |
|
|
Integrated Informatics Platform for Glycans
The type of data sets needed to fully define structure-function relationships of glycans goes beyond the complex analytical data for structural characterization of glycans. Several international collaborative efforts (Table 2) recognize the need for an integrated glycomics approach and are developing novel resources and technologies to better understand glycan structure-function relationships 33. The data sets and databases developed by these initiatives revolve around three main components that are discussed in detail below (summarized in Table 3): 1) glycan structures, 2) glycan biosynthesis pathways, and 3) glycan-protein interactions.
Table 2. International Glycomics Initiatives
|
Consortium for Functional Glycomics (CFG) (USA) |
www.functionalglycomics.org |
|
Complex Carbohydrates Research Center (CCRC) (USA) |
www.ccrc.uga.edu |
|
Lipid Maps Consortium (USA) |
www.lipidmaps.org |
|
EuroCarbDB (Europe) |
www.eurocarbdb.org |
|
Glycosciences.de (Germany) |
www.glycosciences.de |
|
UK Glycochips Consortium (UK) |
www.glycoarrays.org.uk |
|
Centre for Glycobiology (Israel) |
www.bgu.ac.il/glyco/ |
|
Human Disease Glycomics/Proteome Initiative (HGPI) (Japan) |
www.hgpi.jp |
|
Kyoto Encyclopedia of Genes and Genomes (KEGG) Glycan (Japan) |
www.genome.jp/kegg/glycan/ |
Table 3. Open access online web-based resources for glycomics
|
Glycan Structures and Analytical Data |
|
|
|
CFG Glycan Structures DB |
www.functionalglycomics.org/glycomics/molecule/jsp/carbohydrate/carbMoleculeHome.jsp |
Glycans from CarbBank, synthesized by CFG, and glycan topologies used for annotation of CFG MALDI-MS data. |
|
Glycosciences Glycan Structures DB |
www.glycosciences.de/sweetdb/index.php |
Glycans from CarbBank and crystal structures of glycoproteins or glycan-protein complexes in Protein Data Bank. |
|
|
||
|
Glycome-DB |
www.glycome-db.org/About.action |
Compilation of glycan structures from different databases translated using a standardized Glyco-CT XML format. |
|
KEGG Glycan Structures DB |
www.genome.jp/kegg/glycan/ |
Includes glycans from CarbBank, recently published in literature, and from reaction pathways of glycan biosynthesis. |
|
|
||
|
|
||
|
Bacterial Carbohydrate Structures Database |
www.glyco.ac.ru/bcsdb/start.shtml |
Most updated database on bacterial glycan structures and is updated constantly with new structures published in literature. |
|
|
||
|
|
||
|
Glycobase |
glycobase.univ-lille1.fr/base/ |
Glycan structures found in literature derived from selected biological species. |
|
Glycoconjugate DB |
akashia.sci.hokudai.ac.jp/www.functionalglycomics.org/glycomics/publicdata/glycoprofiling.jsp |
Glycan structures in the Protein Data Bank. |
|
CFG |
Raw and annotated MALDI-MS glycan profiles of mouse and human cells and tissues, and integration with |
|
|
MALDI-MS |
||
|
Glycan Profile of Cells and Tissues |
|
|
|
SUGABASE NMR DB |
boc.chem.uu.nl/sugabase/sugabase.html |
Glycan NMR database that is integrated with CarbBank. |
|
CCRC NMR and MS Data |
www.ccrc.uga.edu/specdb/specdbframe.html |
Proton NMR spectra of Xlyoglucans and GC-EIMS of methylated alditol acetates. |
|
Glycan Binding Proteins (GBPs) and Glycan-Protein Interactions |
||
|
CFG GBP DB |
www.functionalglycomics.org/glycomics/molecule/jsp/gbpMolecule-home.jsp |
An integrated portal to mammalian GBPs with links to public databases and CFG data. |
|
|
||
|
Thorkild’s Lectin Page |
plab.ku.dk/tcbh/lectin-links.htm |
Contains useful definitions and updates on discussions/meetings pertaining to lectins. |
|
|
||
|
CFG Glycan Array Screening Data |
www.functionalglycomics.org/glycomics/publicdata/primaryscreen.jsp |
Data on protein binding to each glycan in the array with links to their structures in CFG glycan structures DB. |
|
|
||
|
Lectines 3D |
www.cermav.cnrs.fr/lectines/ |
Links to crystal structures of lectins in PDB organized based on algal, fungal, bacterial, viral, plant, and animal lectins. |
|
GlycoEpitope DB |
www.glyco.is.ritsumei.ac.jp/epitope/ |
Database of glycan epitopes for antibodies and the antibodies that recognize these epitopes. |
|
|
|
|
|
SugarBindDB |
sugarbinddb.mitre.org/ |
Database of known glycan ligands for pathogens. |
|
Glycan Biosynthesis and Biology |
|
|
|
CFG GT DB |
www.functionalglycomics.org/glycomics/molecule/jsp/glycoEnzyme/geMolecule.jsp |
Mammalian glycosylation pathways and an integrated portal to information and data on glycosyltransferases. |
|
KEGG GT DB |
www.genome.jp/kegg-bin/get_htext7ko01003.keg |
Database and tools pertaining to glycosylation pathways and glycosyltransferases. |
|
Carbohydrate Active Enzymes (CAZy) DB |
www.cazy.org/fam/acc_GT.html |
Glycosyltransferases from different species classified according to families depending on their substrate specificity. |
|
CFG Gene Expression Data |
www.functionalglycomics.org/glycomics/publicdata/microarray.jsp |
Expression of mouse and human glycan biosynthesis enzymes and GBPs in various cells under different conditions. |
|
CFG Phenotyping Knockout Mice Data |
www.functionalglycomics.org/glycomics/publicdata/phenotyping.jsp |
Hematology, histology, immunology, and metabolism phenotype analysis of knockout mice in GBPs or GTs. |
Glycan structures database
The common goal of all the major glycomics initiatives is the development of a database of glycan structures in parallel with the analytical data sets (described above). The common source for the initial set of glycan structures to seed different glycan databases was the CarbBank database (34). This database was discontinued in the 1990s because of a lack of resources that was required to sustain its development and ensure the quality of glycan structures based on extensive curation. The diversity in the sugar building blocks of glycans, the link between the sugars and two-dimensional topology (degree and extent of branching), offers challenges in the development of standardized data formats to represent glycan structures across different databases. Because of this obstacle, independent data formats were developed in the various international initiatives to encode glycan structures in databases (35). An important issue develops from the ambiguities in glycan structure that result from incomplete assignment of their exact structure. These ambiguities include unspecified links, anomeric configurations, and uncertainties in the connectivity of a specific sugar (typically terminal sugar) or a set of sugars to a particular two-dimensional glycan topology. Consequently, different databases employ disparate mechanisms to manage these ambiguities; they represent ambiguities as they are or deconvolute them into a set of likely exact structures. An important feature of the glycan structures databases is the tools to query the database using parameters such as monosaccharide composition, substructure, and mass. Different algorithms have been developed to search, score, and compare glycan structures using tree matching methods, scoring matrices, and dynamic programming tools (36-38). The interpretation of the search results from these tools is still demanding primarily because of the lack of structured vocabularies to enable a standardized definition of glycan structures (across different databases) in terms of their structural determination, classification based on synthesis, and biological source. Querying different glycan structures databases using the same query parameters often gives dramatically different results in this context.
Glycan biosynthesis pathways
The Consortium for Functional Glycomics (CFG) has led a collaboration with Japanese and European initiatives to develop a comprehensive annotation of the enzymes involved in glycan biosynthesis. Currently, these annotations are disseminated using glycosylation pathway interfaces and glycosyltransferase databases of the CFG, Kyoto Encyclopedia of Genes and Genomes (KEGG) glycan, Complex Carbohydrates Research Center (CCRC), and CAZy. The CFG has developed customized DNA microarrays that appropriately represent glycan biosynthesis and binding protein genes to study their expression pattern (39). Hundred of samples representing various tissues and cell types have been analyzed on the CFG glyco-gene microarrays. The focus of experiments includes the analysis of different tumor cell lines, cell types, or tissues from glycosyltransferase or glycan binding protein knockout mice strains, cells under mechanical stress such as chondrocytes, and so forth. The CFG recently has been generating both gene expression and MALDI-MS glycan profile data sets simultaneously for different cells and tissues (40). Algorithms to predict glycan structures based on expression profile of glycosyltransferases are in development (41). The integration of these data sets in the CFG databases thus enables the application of these algorithms to correlate the glycan structure profile with the expression of the glycan biosynthesis enzymes in a specific cell line. Another valuable resource for quantitative analysis of gene expression is the quantitative real-time PCR analysis of glycan-related genes by the CCRC.
In addition to the genome-level analysis of glycan biosynthesis pathways, the CFG is also working to generate and phenotype transgenic mouse lines that represent knockouts later stage fucosyl- and sialyl-transferases. These transgenic mice are subject to a battery of phenotype analysis studies viz: 1) hematology and coagulation chemistry, 2) histological staining of tissues, 3) immunology assays such as FACS, Ig levels, measuring B- and T-cell proliferation during induction with various agents and cytokines, and 4) various metabolism and behavioral tests. These studies have generated volumes of new data that offer numerous parameters to quantify the distinct phenotypic abnormalities in these mice.
Glycan-protein interactions
Understanding specificity of glycan-protein interactions that govern a biological process provides the right context to define glycan sequence and to comprehend glycan structure-function relationships. The binding site (or carbohydrate recognition domain; CRD) on glycan binding proteins (GBPs) typically accommodates monohexasaccharide glycan ligand motifs (1, 2, 42). The monosaccharide identity, anomeric configuration, and link, as well as the overall structural topology of the glycan ligand critically influence binding specificity to a given protein. The majority of the physiological glycan-GBP interactions are multivalent and involve binding of an ensemble of glycan motifs to multimeric CRDs formed by association of GBPs (Fig. 2). This multivalency plays an additional critical role in governing the biological specificity of glycan-protein interactions. In the case of GAGs, chain length, chain conformation (provides a distinct structural topology), and sulfation pattern are critical determinants of the biological specificity mediated by GAG-protein interactions (Fig. 2).
Advances in chemical and chemo-enzymatic synthesis have resulted in the synthesis of hundreds of glycan structures that capture the diversity of the glycans present at the cell surface (43-47). Glycan arrays that contain these synthetic glycans are becoming widely used resources to gain a rapid screen for gly- can binding of a given sample (43, 48, 49). The presentation of the glycans on these arrays reasonably mimics the physiological presentation of these glycans on the cell surface. Diverse samples including mammalian GBPs, plant lectins, pathogen surface proteins and whole pathogens, antibodies, and cells have been screened on CFG glycan arrays. On several occasions, the data generated from these analyses have uncovered novel glycan recognition motifs for these diverse range of samples analyzed. The CFG and other groups have also developed specific databases for GBPs and other lectins (from plants and bacteria) that capture the sequence, three-dimensional structure, known glycan binding specificities, and other information from the proteins (Table 3).

Figure 2. Multivalent glycan-protein interactions. Shown on the left is a schematic of multivalent interactions between branched N-linked and O-linked glycans on glycoproteins with CRDs glycan binding proteins. The binding specificity is critically governed by the chemical structure and three-dimensional topology of the glycan ligand motif and the multivalency. Given that the glycan ligand motif can be present on the same or different glycoproteins, it is important to characterize the ensemble of glycans isolated from a given cell. Shown on the right is a schematic of GAG-protein interactions. GAGs are typically involved in facilitating the assembly of protein-protein complexes, enzyme-inhibitor complexes, and ligand-receptor complexes on the cell surface and thus play important roles in extracellular signaling. The schematic shows an example of HS GAG (H-glucosamine; I-iduronic acid) wherein the chain length, chain conformation, and sulfation pattern governs whether an active or inactive protein-protein complex is assembled.
Data integration for glycomics
Clearly, a need to cut across multiple data sets to understand the structure-function relationships of glycans is apparent. A critical component that permits this process is the bioinformatics platform used to store, integrate, and process the information generated by the above methods and to disseminate this information in a meaningful fashion via the Internet to the scientific community worldwide (35, 50). To capture complex relationships between diverse data, it is necessary to develop an object-based relational database. Three primary objects are found in glycomics data sets: GBPs, glycan biosynthetic enzymes, and the glycan structures. The different methodologies that generate data sets are organized into secondary and other levels of objects with defined interrelationships and relationships to the primary objects. The overall scheme for data integration in the CFG databases is shown as an example (Fig. 3). The key identifiers to uniquely define the primary objects include the glycan structure represented in the internal database format, primary sequences (or accession numbers such as GenBank and SwissProt) of the GBPs, and glycan biosynthetic enzymes. The relationship of primary to secondary data objects is established using object-based keys such as sample, protocols, and data keys. Unlike in the case of the primary objects, it is challenging to define standardized criteria that can determine the uniqueness of the secondary objects (such as sample information) because of the lack of standardized ontologies to describe sample and protocol information for glycomics. To address this challenge, a key step would require outlining metadata standards for obtaining various glycomics data sets, including mass-spectrometric glycan analysis data and glycan-protein interaction data. It is important, for example, to capture mass spectrometer instrument settings, internal standards, and other experimental conditions used to characterize glycans from a given sample. The blueprint of the object-based relational database is a data model that captures data definitions and interrelationships between the data sets using the specific object-based identifiers, which is quite complex for glycomics databases. It is important, therefore, to develop a software architecture that keeps this complexity hidden from the user during the actual data acquisition and dissemination. The three-tier software architecture containing a back-end relational database to store the data and annotate relationships, a middleware application layer that communicates between the database and the user interface, and the top layer that comprises the user interfaces to the database is best suited for this purpose. This software architecture facilitates the easy deposit of data into the database, which is organized automatically into the relational tables by the middleware application layer.
Central to this data integration is an ability to link orthogonal data sets derived from identical, or similar, samples. For example, the gene expression profile of a specific tissue or cell line isolated from a given strain of transgenic mice needs to be automatically associated in the database with orthogonal information such as glycan profile, histological staining, and immunological profile from a similar or identical sample. Such integration would permit researchers to cut across multiple data sets and begin posing questions such as “Does the expression of glycosyltransferase correlate with glycan profile of that tissue?” or “Can the pathological analysis of the tissue be explained on the basis of gene expression profile?” The bioinformatics platform ultimately needs to support computational tools that perform data mining analysis on the large-scale glycomics data sets. The prediction of glycan structures based on gene expression profiles of glycan biosynthetic enzymes (41) is now enabled because of the user-friendly access to diverse data sets via relational databases.
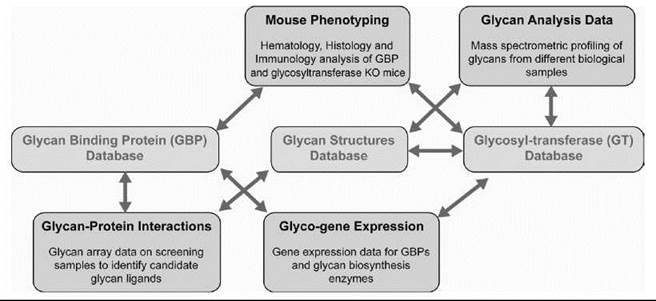
Figure 3. Data integration in glycomics. An example of integrating the diverse data sets generated by CFG to the three primary databases viz glycan structures, GBP, and GT is shown. The arrows indicate the connectivity between the objects using the primary identifiers for genes, proteins, glycan structures, and samples analyzed. This integration enables seamless navigation across these data sets. For example, starting from a glycosylation pathway, it is possible to navigate to a specific glycan structure whose biosynthesis involves a particular enzyme. If a glycan ligand motif present on the glycan array is a part of this glycan structure, it is possible to determine which of the samples analyzed on the array gave good binding signals to this motif. If any of the samples analyzed are mammalian GBPs, then it is possible to access an entire portal of information on that particular GBP.
Significance and Future Directions
Glycomics is an emerging field that adds new dimensions to both technology development and obtaining an understanding of fundamental biological processes in this age of postgenomics. The two fundamental questions advancing glycomics are as follows: 1) How do we define glycan diversity in the context of biosynthesis and specificity in modulating a biological process and 2) how do we define specificity in the context of multivalent glycan-protein interactions? It is important to consider these questions in light of the numerous efforts underway to discover novel glycan-based biomarkers and therapeutic targets (51-54). In the case of biomarker discovery, two distinct approaches are found. The glycobiology approach begins with the discovery of a specific glycan ligand motif specifically involved in a biologically important glycan-protein interaction. The use of this motif as a biomarker then is investigated by studying the expression of this motif as a part of glycan structure or glycan biosynthesis enzymes that generate this motif in different biological samples. The other approach is more of a high-throughput analytical approach that relies on the various analytical and informatics tools to characterize glycan structures in different samples and to identify unique markers based on differential expression of specific structures in a given sample. The generation of diverse data sets and their integration via an informatics platform is critical to both approaches. From an informatics perspective, the key issues that need to be addressed are as follows: 1) developing standardization and quality control of the data, 2) developing consistent format for representation and transfer of complex glycan structures across different databases, and 3) developing robust ontologies and structured vocabulary for glycomics (55).
A significant portion of the high-throughput data sets on glycan analysis and glycan-protein interactions are fairly recent, and therefore a critical need is found to standardize the generation of these data so they can be compared across data sets generated by different initiatives (35). Also, awareness is increasing of the need to develop data exchange formats, such as XML, for consistent description of glycan structures and glycomics data sets across different large-scale glycomics initiatives (19). Leading glycomics initiatives recently agreed to adopt the Glyde-II XML format (52). The next step toward standardization of glycan structure databases is to establish the standards for incorporating glycan structures into a database to develop the glycan database into an international resource similar to GenBank and SwissProt. Collaborative discussions between the large-scale glycomics initiatives to address these issues are notable steps toward advancing glycomics.
Acknowledgments
This work was supported by the National Institute of General Medical Sciences Glue Grant U54 GM62116. The authors thank the CFG members, the members of Bioinformatics Core (B) of CFG, for the work described in this review, and V. Sasisekharan for critical reading of the manuscript.
References
1. Varki A, Cummings R, Esko JD, Freeze H, Hart GW, Marth J. Essentials of Glycobiology. 1999. Cold Spring Harber Laboratory Press, New York.
2. Taylor ME, Drickamer K. Introduction to Glycobiology. 2003. Oxford University Press, Oxford, UK.
3. Sasisekharan R, Raman R, Prabhakar V. Glycomics approach to structure-function relationships of glycosaminoglycans. Annu. Rev. Biomed. Eng. 2006; 8:181-231.
4. Arnold JN, Wormald MR, Sim RB, Rudd PM, Dwek RA. The impact of glycosylation on the biological function and structure of human immunoglobulins. Annu. Rev. Immunol. 2007; 25:21-50.
5. Bishop JR, Schuksz M, Esko JD. Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature 2007; 446:1030-1037.
6. Crocker PR, Paulson JC, Varki A. Siglecs and their roles in the immune system. Nat. Rev. Immunol. 2007; 7:255-266.
7. Dube DH, Bertozzi CR. Glycans in cancer and inflammation—potential for therapeutics and diagnostics. Nat. Rev. Drug Discov. 2005; 4:477-488.
8. Haltiwanger RS, Lowe JB. Role of glycosylation in development. Annu. Rev. Biochem. 2004; 73:491-537.
9. Collins BE, Paulson JC. Cell surface biology mediated by low affinity multivalent protein-glycan interactions. Curr. Opin. Chem. Biol. 2004; 8:617-625.
10. Raman R, Sasisekharan R. Cooperativity in glycan-protein interactions. Chem. Biol. 2007; 14:873-874.
11. Kaji H, Saito H, Yamauchi Y, Shinkawa T, Taoka M, Hirabayashi J, Kasai K, Takahashi N, Isobe T. Lectin affinity capture, isotope-coded tagging and mass spectrometry to identify N-linked glycoproteins. Nat. Biotechnol. 2003; 21:667-672.
12. Karlsson NG, Wilson NL, Wirth HJ, Dawes P, Joshi H, Packer NH. Negative ion graphitised carbon nano-liquid chromatography/mass spectrometry increases sensitivity for glycoprotein oligosaccharide analysis. Rapid Commun. Mass Spectrom. 2004; 18:2282-2292.
13. Harvey DJ. Analysis of carbohydrates and glycoconjugates by matrix-assisted laser desorption/ionization mass spectrometry: An update covering the period 1999-2000. Mass Spectrom Rev 2006; 25:595-662.
14. Wada Y, Azadi P, Costello CE, Dell A, Dwek RA, Geyer H, Geyer R, Kakehi K, Karlsson NG, Kato K, Kawasaki N, Khoo KH, Kim S, et al. Comparison of the methods for profiling glycoprotein glycans-HUPO Human Disease Glycomics/Proteome Initiative multi-institutional study. Glycobiology 2007; 17:411-422.
15. Zaia J. Mass spectrometry of oligosaccharides. Mass Spectrom. Rev. 2004; 23:161-227.
16. Park Y, Lebrilla CB. Application of Fourier transform ion cyclotron resonance mass spectrometry to oligosaccharides. Mass Spectrom. Rev. 2005; 24:232-264.
17. Cooper CA, Gasteiger E, Packer NH. GlycoMod—a software tool for determining glycosylation compositions from mass spectrometric data. Proteomics 2001; 1:340-349.
18. Goldberg D, Sutton-Smith M, Paulson J, Dell A. Automatic annotation of matrix-assisted laser desorption/ionization N-glycan spectra. Proteomics 2005; 5:865-875.
19. Sahoo SS, Thomas C, Sheth A, Henson C, York WS. GLYDE-an expressive XML standard for the representation of glycan structure. Carbohydr. Res. 2005; 340:2802-2807.
20. Maass K, Ranzinger R, Geyer H, von der Lieth CW, Geyer R. “Glyco-peakfinder”-de novo composition analysis of glycoconjugates. Proteomics 2007; 7:4435-4444.
21. Ceroni A, Dell A, Haslam SM. The GlycanBuilder: a fast, intuitive and flexible software tool for building and displaying glycan structures. Source Code Biol. Med. 2007; 2:3.
22. Josh HJ, Harrison MJ, Schulz BL, Cooper CA, Packer NH, Karlsson NG. Development of a mass fingerprinting tool for automated interpretation of oligosaccharide fragmentation data. Proteomics 2004; 4:1650-64.
23. Gaucher SP, Morrow J, Leary JA. STAT: a saccharide topology analysis tool used in combination with tandem mass spectrometry. Anal Chem 2000; 72:2331-2336.
24. Ethier M, Saba JA, Ens W, Standing KG, Perreault H. Automated structural assignment of derivatized complex N-linked oligosaccharides from tandem mass spectra. Rapid Commun. Mass Spectrom. 2002; 16:1743-1754.
25. Lapadula AJ, Hatcher PJ, Hanneman AJ, Ashline DJ, Zhang H, Reinhold VN. Congruent strategies for carbohydrate sequencing. 3. OSCAR: an algorithm for assigning oligosaccharide topology from MSn data. Anal. Chem. 2005; 77:6271-6279.
26. Rudd PM, Colominas C, Royle L, Murphy N, Hart E, Merry AH, Hebestreit HF, Dwek RA. A high-performance liquid chromatography based strategy for rapid, sensitive sequencing of N-linked oligosaccharide modifications to proteins in sodium dodecyl sulphate polyacrylamide electrophoresis gel bands. Proteomics 2001; 1:285-294.
27. Venkataraman G, Shriver Z, Raman R, Sasisekharan R. Sequencing complex polysaccharides. Science 1999; 286:537-542.
28. Guerrini M, Raman R, Venkataraman G, Torri G, Sasisekharan R, Casu B. A novel computational approach to integrate NMR spectroscopy and capillary electrophoresis for strucsture assignment of heparin and heparan sulfate oligosaccharides. Glycobiology 2002; 12:713-719.
29. Tang H, Mechref Y, Novotny MV. Automated interpretation of MS/MS spectra of oligosaccharides. Bioinformatics 2005; 21(Suppl 1):i431-i439.
30. Zhang H, Singh S, Reinhold VN. Congruent strategies for carbohydrate sequencing. 2. FragLib: an MSn spectral library. Anal. Chem. 2005; 77:6263-6270.
31. Royle L, Mattu TS, Hart E, Langridge JI, Merry AH, Murphy N, Harvey DJ, Dwek RA, Rudd PM. An analytical and structural database provides a strategy for sequencing O-glycans from microgram quantities of glycoproteins. Anal. Biochem. 2002; 304:70-90.
32. Manzi AE, Norgard-Sumnicht K, Argade S, Marth JD, van Halbeek H, Varki A. Exploring the glycan repertoire of genetically modified mice by isolation and profiling of the major glycan classes and nano-NMR analysis of glycan mixtures. Glycobiology 2000; 10:669-689.
33. Raman R, Raguram S, Venkataraman G, Paulson JC, Sasisekharan R. Glycomics: an integrated systems approach to structure-function relationships of glycans. Nat. Methods 2005; 2:817-824.
34. Doubet S, Albersheim P. CarbBank. Glycobiology 1992; 2:505.
35. von der Lieth CW, Bohne-Lang A, Lohmann KK Frank M. Bioinformatics for glycomics: status, methods, requirements and perspectives. Brief Bioinform. 2004; 5:164-178.
36. Aoki KF, Mamitsuka H, Akutsu T, Kanehisa M. A score matrix to reveal the hidden links in glycans. Bioinformatics 2005; 21:1457-1463.
37. Aoki KF, Yamaguchi A, Ueda N, Akutsu T, Mamitsuka H, Goto S, Kanehisa M. KCaM (KEGG Carbohydrate Matcher): a software tool for analyzing the structures of carbohydrate sugar chains. Nucleic Acids Res. 2004; 32:W267-272.
38. Aoki-Kinoshita KF, Ueda N, Mamitsuka H, Kanehisa M. ProfilePSTMM: capturing tree-structure motifs in carbohydrate sugar chains. Bioinformatics 2006; 22:e25-34.
39. Comelli EM, Amado M, Head SR, Paulson JC. Custom microarray for glycobiologists: considerations for glycosyltransferase gene expression profiling. Biochem. Soc. Symp. 2002; 135-142.
40. Comelli EM, Head SR, Gilmartin T, Whisenant T, Haslam S. North, SJ, Wong NK, Kudo T, Narimatsu H, Esko JD, Drick- amer K, Dell A, Paulson JC. A focused microarray approach to functional glycomics: transcriptional regulation of the glycome. Glycobiology 2006; 16:117-131.
41. Kawano S, Hashimoto K, Miyama T, Goto S, Kanehisa M. Prediction of glycan structures from gene expression data based on glycosyltransferase reactions. Bioinformatics 2005; 21:3976-3982.
42. Sharon N, Lis H. Lectins. 2004. Kluwer Academic Publishers, Boston, MA.
43. Blixt O, Head S, Mondala T, Scanlan C, Huflejt ME, Alvarez R, Bryan MC, Fazio F, Calarese D, Stevens J, Razi N, Stevens DJ, Skehel JJ, van Die I, Burton DR, Wilson IA, Cummings R, Bovin N, Wong CH, Paulson JC. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:17033-17038.
44. Hanson S, Best M, Bryan MC, Wong CH. Chemoenzymatic synthesis of oligosaccharides and glycoproteins. Trends Biochem. Sci. 2004; 29:656-663.
45. Plante OJ, Palmacci ER, Seeberger PH. Automated solid-phase synthesis of oligosaccharides. Science 2001; 291:1523-1527.
46. Seeberger PH, Werz DB. Synthesis and medical applications of oligosaccharides. Nature 2007; 446:1046-1051.
47. Werz DB, Ranzinger R, Herget S, Adibekian A, von der Lieth CW, Seeberger PH. Exploring the structural diversity of mammalian carbohydrates (“glycospace”) by statistical databank analysis. ACS Chem. Biol. 2007; 2:685-691.
48. Feizi T, Fazio F, Chai W, Wong CH. Carbohydrate microarrays—a new set of technologies at the frontiers of glycomics. Curr. Opin. Struct. Biol. 2003; 13:637-645.
49. Galanina OE, Mecklenburg M, Nifantiev NE, Pazynina GV, Bovin NV. GlycoChip: multiarray for the study of carbohydrate-binding proteins. Lab Chip 2003; 3:260-265.
50. Raman R, Venkataraman M, Ramakrishnan S, Lang W, Raguram S, Sasisekharan R. Advancing glycomics: implementation strategies at the consortium for functional glycomics. Glycobiology 2006; 16:82R-90R.
51. Aoki-Kinoshita KF, Kanehisa M. Bioinformatics approaches in glycomics and drug discovery. Curr. Opin. Mol. Ther. 2006; 8:514-520.
52. Packer NH, von der Lieth CW Aoki-Kinoshita KF, Lebrilla CB, Paulson JC, Raman R, Rudd P, Sasisekharan R, Taniguchi N, York WS. Frontiers in glycomics: Bioinformatics and biomarkers in disease An NIH White Paper prepared from discussions by the focus groups at a workshop on the NIH campus, Bethesda MD (September 11-13, 2006). Proteomics 2007; 8:8-20.
53. Shriver Z, Raguram S, Sasisekharan R. Glycomics: a pathway to a class of new and improved therapeutics. Nat. Rev. Drug Discov. 2004; 3:863-873.
54. von der Lieth CW, Lutteke T, Frank M. The role of informatics in glycobiology research with special emphasis on automatic interpretation of MS spectra. Biochim. Biophys. Acta 2006; 1760:568-577.
55. Sahoo SS, Thomas C, Sheth A, York WS, Tartir S. Knowledge modeling and its application in life sciences: a tale of two ontologies in 2006. Proceedings of the 15th International Conference on World Wide Web, Edinburgh, Scotland.