MCAT Biology and Biochemistry: New for MCAT 2015 (2014)
Chapter 6. Microbiology
A milestone in microbiology was the demonstration by Louis Pasteur in 1861 that microbes do not spontaneously arise in boiled broth; they must arrive there by contamination. This put the last nail in the coffin of the idea of spontaneous generation of life. Another major contribution to the golden age of microbiology was the isolation of the bacteria responsible for anthrax in 1876 by a physician named Robert Koch. This and other experiments led to the germ theory of disease, the idea that disease was not caused by bad air (“malaria”), but by microorganisms. In this chapter we will examine three major groups of disease-causing organisms, beginning with the smallest (viruses) and ending with the largest (fungi—which are often not microscopic at all but visible with the naked eye).
6.1 VIRUSES
With the identification of bacteria as the cause of anthrax and other diseases, medical science appeared in the late 1800s to be headed toward explanation of all infectious disease. Researchers soon found however that some infectious agents could not be trapped by passage through filters in the same manner as bacteria. These agents also proved invisible to the light microscope, unlike bacteria. With the advent of electron microscopy, the tiny infectious agents known as viruses were finally visualized. Today, molecular biology has shed great light onto viruses, down to the nucleotide sequence of entire viral genomes. However, viruses such as HIV still remain one of the most serious threats to health, indicating there is still much to learn.
Viruses infect all life forms on earth, including plants, animals, protists, and bacteria. A virus is an obligate intracellular parasite. As such, they are only able (obligated) to reproduce within (intra) cells. While within cells, viruses have some of the attributes of living organisms, such as the ability to reproduce; but outside cells, viruses are without activity. Viruses on their own are unable to perform any of the chemical reactions characteristic of life, such as synthesis of ATP and macromolecules.1 Viruses are not cells or even living organisms. To reproduce, they commandeer the cellular machinery of the host they infect and use it to manufacture copies of themselves. In the final analysis, a virus is nothing more than a package of nucleic acid that says: “Pick me up and reproduce me.” Remember this crucial definition: A virus is an obligate intracellular parasite which relies on host machinery whenever possible. In the following sections we will look at some of the variations on this basic theme.
• Cyanide (an inhibitor of the electron transport chain) is added to a culture of virus-infected mammalian cells. The virus has none of the components of electron transport nor any other proteins that are inhibited by cyanide. Which one of the following best describes the effect of cyanide?2
A) The mammalian cells will die, and all viruses will be destroyed as well, regardless of their stage of development.
B) Mammalian cells are killed, and viral replication halted, but the culture remains infectious.
C) Mammalian cells stop growing, and viral replication is unaffected.
D) Mammalian cells continue to grow, but viral replication is halted.
Viral Structure and Function
The structure of viruses reflects their life cycle. In general, all viruses possess a nucleic acid genome packaged in a protein shell. The exterior protein packaging helps to convey the genome from one cell to infect other cells. Once in a cell, the viral genome directs the production of new copies of the genome and of the protein packaging needed to produce more virus. However, the nature of the genome, the protein packaging, and the viral life cycle vary tremendously between different viruses.
A viral genome may consist of either DNA or RNA that is either single- or double-stranded and is either linear or circular. Viruses utilize virtually every conceivable form of nucleic acid as their genome. However, a given type of virus can have only one type of nucleic acid as its genome, and mature virus does not contain nucleic acid other than its genome.3 [If the ratio of adenine to thymine in a DNA virus is not one to one, what can be said about the genome of this virus?4 A disease agent that is isolated from a human cannot reproduce on its own in cell-free broth but can reproduce in a culture of human cells. In its pure form it possesses both RNA and DNA. Is it possible that the disease agent is a virus?5]
A factor that influences all viral genomes, regardless of the form of the nucleic acid used as genome, is size as a limiting factor. Viruses are much smaller than the hosts they infect, both prokaryotic and eukaryotic. Figure 1 depicts the relative size of a bacteriophage (a virus that infects bacteria) and its host.
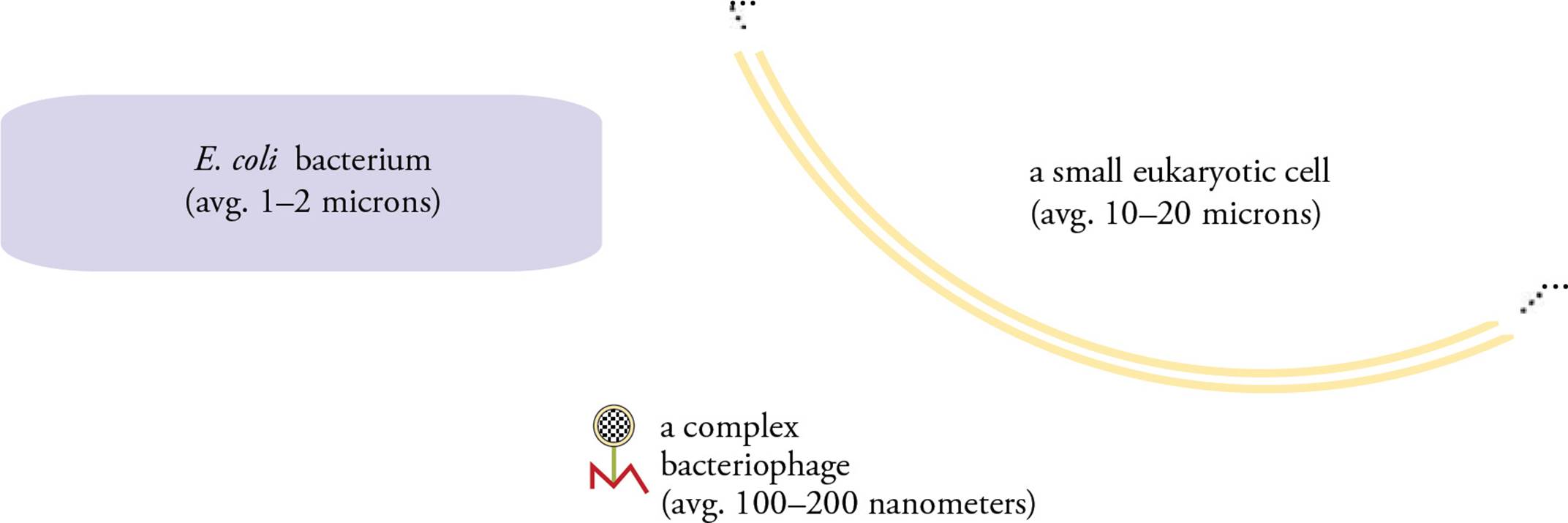
Figure 1 The Relative Size of a Virus
Not only are viruses small, but the exterior protein shell of a virus is typically a rigid structure of fixed size that cannot expand to accommodate a larger genome. [What is the likely result if a viral genome is tripled in size?6] To adapt to this size constraint, viral genomes have evolved to be extremely economical. One adaptation is for the viral genome to carry very few genes and for the virus to rely on host-encoded proteins for transcription, translation, and replication. [How do ribosomes used to translate viral proteins compare to host ribosomes?7] Another adaptation found in viral genomes is the ability to encode more than one protein in a given length of genome. A virus can accomplish this feat by utilizing more than one reading frame within a piece of DNA so that genes may overlap with each other.
• A 1000 base pair region of viral genome is found to encode two polypeptides unrelated in amino acid sequence during infection of eukaryotic cells. If one of these polypeptides is 250 amino acids in length and the other is 300, what is the best explanation for this?8
A) A missense mutation
B) Viruses use a different genetic code than eukaryotes do
C) Overlapping multiple reading frames
D) The polypeptides are splicing variants
Surrounding the viral nucleic acid genome is a protein coat called the capsid. The capsid provides the external morphology that is used to classify viruses. It is made from a repeating pattern of only a few protein building blocks. Helical capsids are rod-shaped, while polyhedral capsids are multiple-sided geometric figures with regular surfaces. Complex viruses may contain a mixture of shapes. For example, the T4 bacteriophage has a helical sheath and a polyhedral head (Figure 2). This virus is commonly used in research; its host is the bacterium E. coli. The genome is located within the capsid head. Other parts of the capsid are used during infection of the host. The tail fibersattach to the surface of the host cell, as does the base plate. The sheath contracts using the energy of stored ATP, injecting the genome into the host. [Why might a bacteriophage inject its DNA, while animal viruses do not?9]
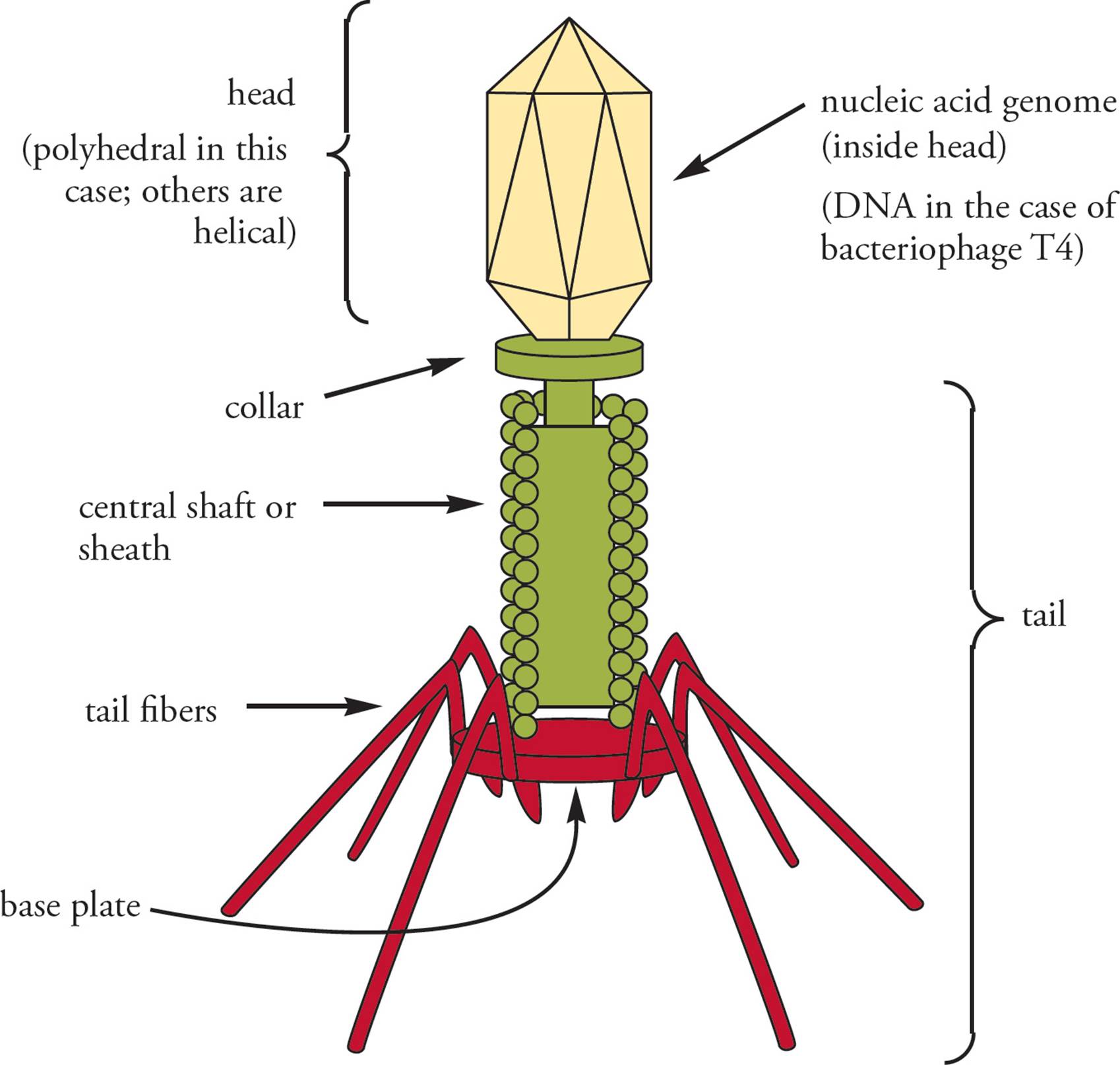
Figure 2 Bacteriophage T4
The most important thing to understand is that the entire viral capsid is composed of protein, while the viral genome is composed of nucleic acid (DNA or RNA). Most viruses are not as structurally complex as the bacteriophage shown in Figure 2. See Figure 3 for more examples.
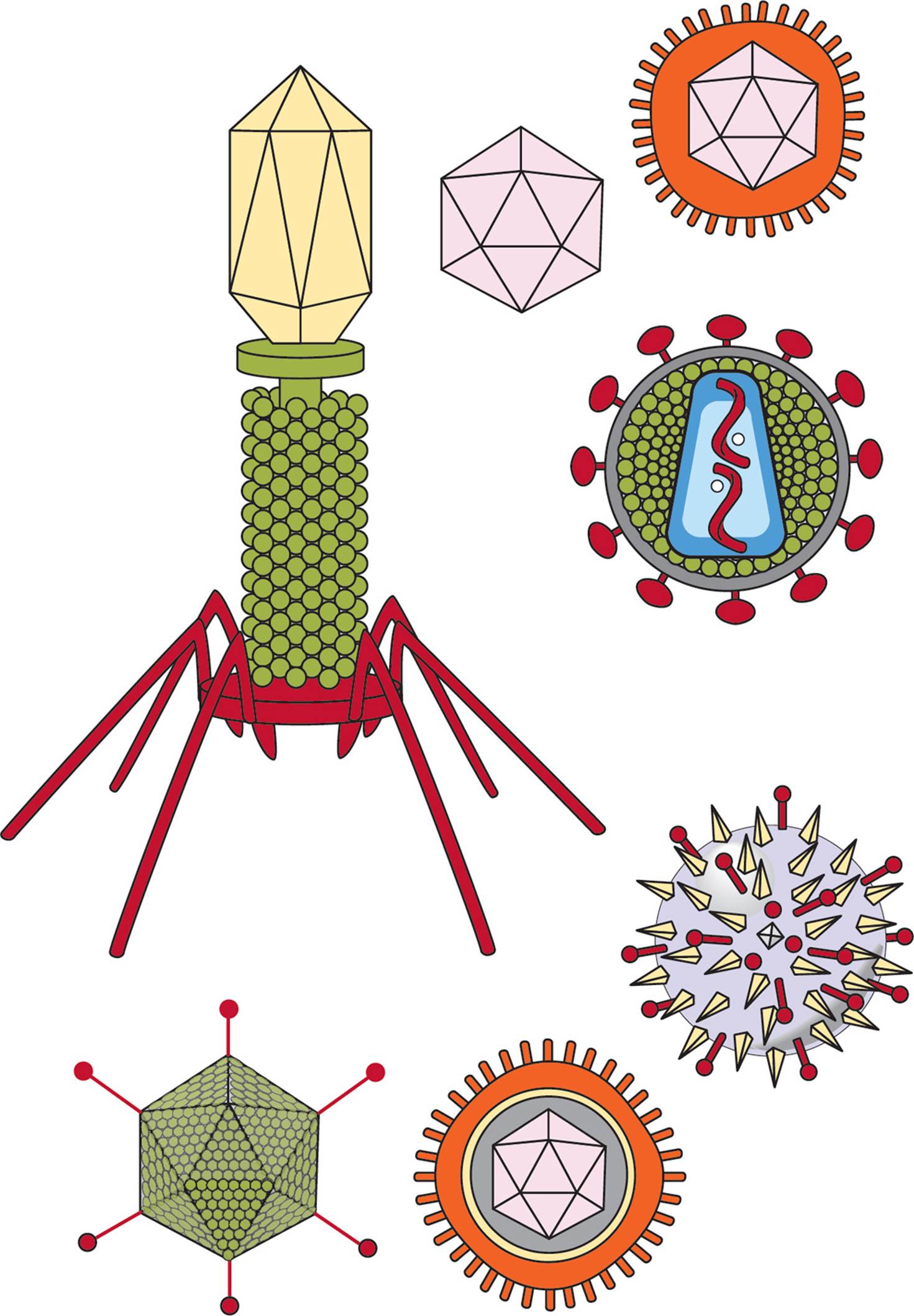
Figure 3 A Variety of Viruses
Many animal viruses also possess an envelope that surrounds the capsid. This is a membrane on the exterior of the virus derived from the membrane of the host cell. It contains phospholipids, proteins, and carbohydrates from the host membrane, in addition to proteins encoded by the viral genome. Enveloped viruses acquire this covering by budding through the host cell membrane. To infect a new host, some enveloped viruses fuse their envelope with the host’s plasma membrane, which leaves the de-enveloped capsid inside the host cell. Viruses which do not have envelopes are called naked viruses. All phages and plant viruses are naked. [Can you imagine why this might be true?10]
Whether enveloped or naked, the surface of a virus determines what host cells it can infect. Viral infection is not a random process, but highly specific. A virus binds to a specific receptor on the cell surface as the first step in infection. After binding, the virus will be internalized, either by fusion with the plasma membrane or by receptor-mediated endocytosis. Only cells with a receptor that matches the virus will become infected, explaining why only specific species or specific cell types are susceptible to infection. The viral surface is also important for recognition by our immune system. [If antibodies to a viral capsid protein are ineffective in blocking infection, what might this indicate about the virus?11]
Bacteriophage Life Cycles
Since viruses lack the ability to produce energy and replicate on their own, they use the machinery of the cell they infect to carry out these processes. The viral genome contains genes that redirect the infected cell to produce viral products. The first step is binding to the exterior of a bacterial cell in a process termed attachment or adsorption. The next step is injection of the viral genome into the host cell in a process termed penetration or eclipse. It is called “eclipse” because the capsid remains on the outer surface of the bacterium while the genome disappears into the cell, removing infectious virus from the media. From this point forward a phage follows one of two different paths: It enters either the lytic cycle or the lysogenic cycle.
The Lytic Cycle of Phages
As soon as the phage genome has entered the host cell, host polymerases and/or ribosomes begin to rapidly transcribe and translate it. One of the first viral gene products made is sometimes an enzyme called hydrolase, a hydrolytic enzyme that degrades the entire host genome. (Hydrolase is an example of an early gene; one of a group of genes that are expressed immediately after infection and which includes any special enzymes required to express viral genes.) Then multiple copies of the phage genome are produced (using the dNTPs resulting from degradation of the host genome), as well as an abundance of capsid proteins. Next, each new capsid automatically assembles itself around a new genome. Finally, an enzyme called lysozyme is produced. An example of a late gene, lysozyme is also present in human tears and saliva. It destroys the bacterial cell wall. Because osmotic pressure is no longer counteracted by the protection of the cell wall, the host bacterium bursts (“lyses,” hence the name lytic), releasing about 100 progeny viruses, which can begin another round of the cycle (see Figure 4). [If lysozyme were an early gene, would this be advantageous to the virus?12]
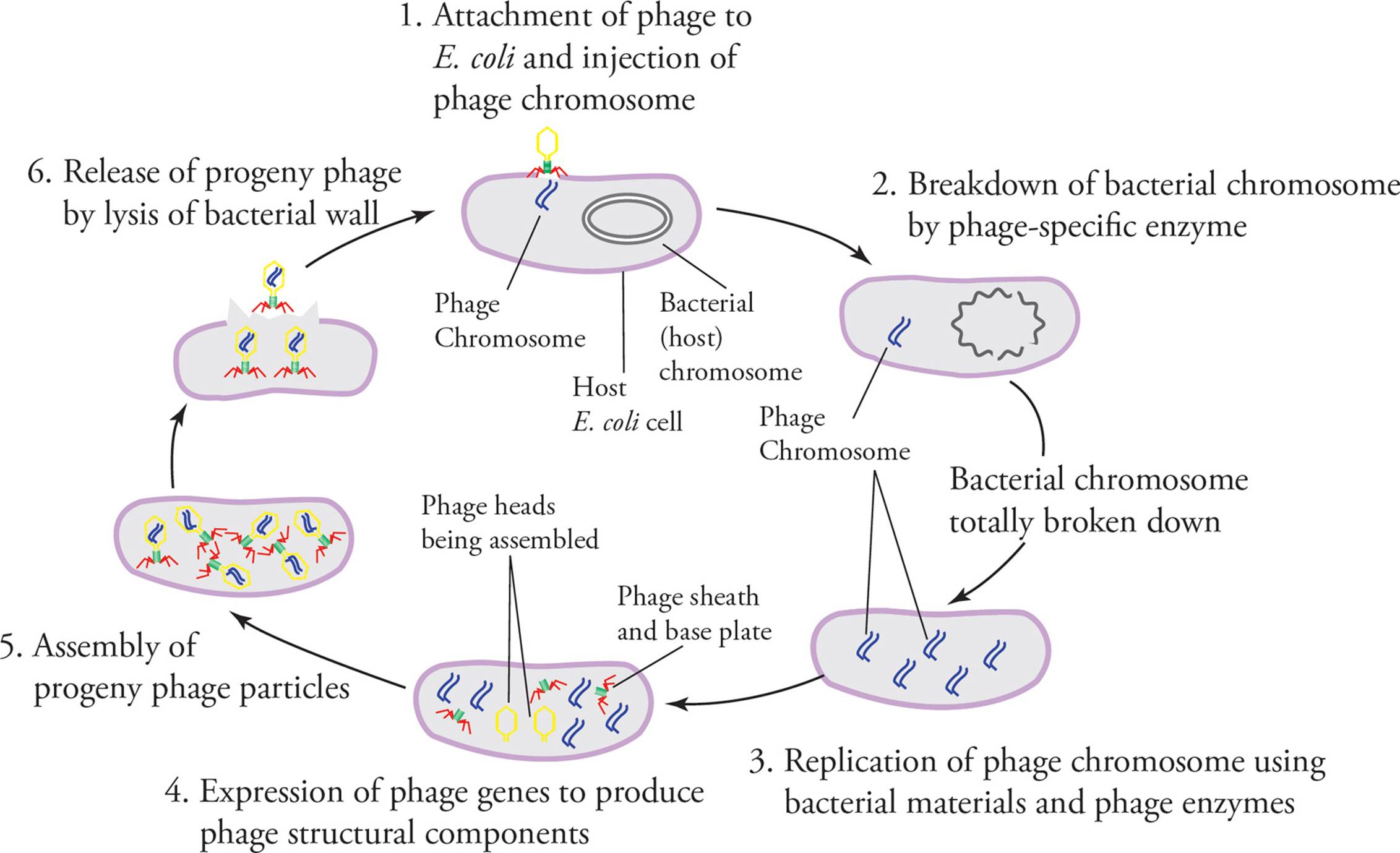
Figure 4 The Lytic Cycle
• When phage are first added to a bacterial culture, the number of infective viruses initially decreases before it later increases. Why does this occur?13
• Bacteria cultured in the presence of 35S-labeled cysteine and 32P-labeled phosphates are infected with phage T4. When phage from this culture are used to infect a new nonradiolabeled bacterial culture, which of the isotopes will be found in the interior of the newly-infected bacteria?14
• A bacteriophage with an important capsid gene deleted infects the same cell as another virus with a normal copy of the same gene. At the time of host-cell lysis:15
A) all released viruses will be capable of infecting new hosts, but only some of these new infections will give rise to phage capable of infecting new hosts.
B) no infective viruses will be released.
C) each individual virus that is released will produce a mixture of infective and noninfective viruses in subsequent infections.
D) only normal viruses will be released.
The Lysogenic Cycle of Phages
The lytic cycle is an efficient way for a virus to rapidly increase its numbers. It presents a problem though: All host cells are destroyed. This is an evolutionary disadvantage. Some viruses are cleverer: They enter the lysogenic cycle. Upon infection, the phage genome is incorporated into the bacterial genome and is now referred to as a prophage; the host is now called a lysogen (Figure 5). The prophage is silent; its genes are not expressed, and viral progeny are not produced. This dormancy is due to the fact that transcription of phage genes is blocked by a phage-encoded repressor protein that binds to specific DNA elements in phage promoters (operators). The cleverness of the lysogenic cycle lies in the fact that every time the host cell reproduces itself, the prophage is reproduced too. Eventually, the prophage becomes activated. It now removes itself from the host genome (in a process called excision) and enters the lytic cycle.
One potential consequence of the lysogenic cycle is that when the viral genome activates, excising itself from the host genome, it may take part of the host genome along with it. When the virus replicates, the small piece of host genome will be replicated and packaged with the viral genome. In subsequent infections, the virus will integrate the “stolen” host DNA along with its own genome into the new host’s genome. The presence of the new DNA will become evident if it codes for a trait that the newly-infected host did not previously possess, such as the ability to metabolize galactose. This process is called transduction. [Why would a bacterial gene, carried with a virus and integrated with viral genes into a new bacterial genome, not be repressed along with the viral genes during lysogeny?16]
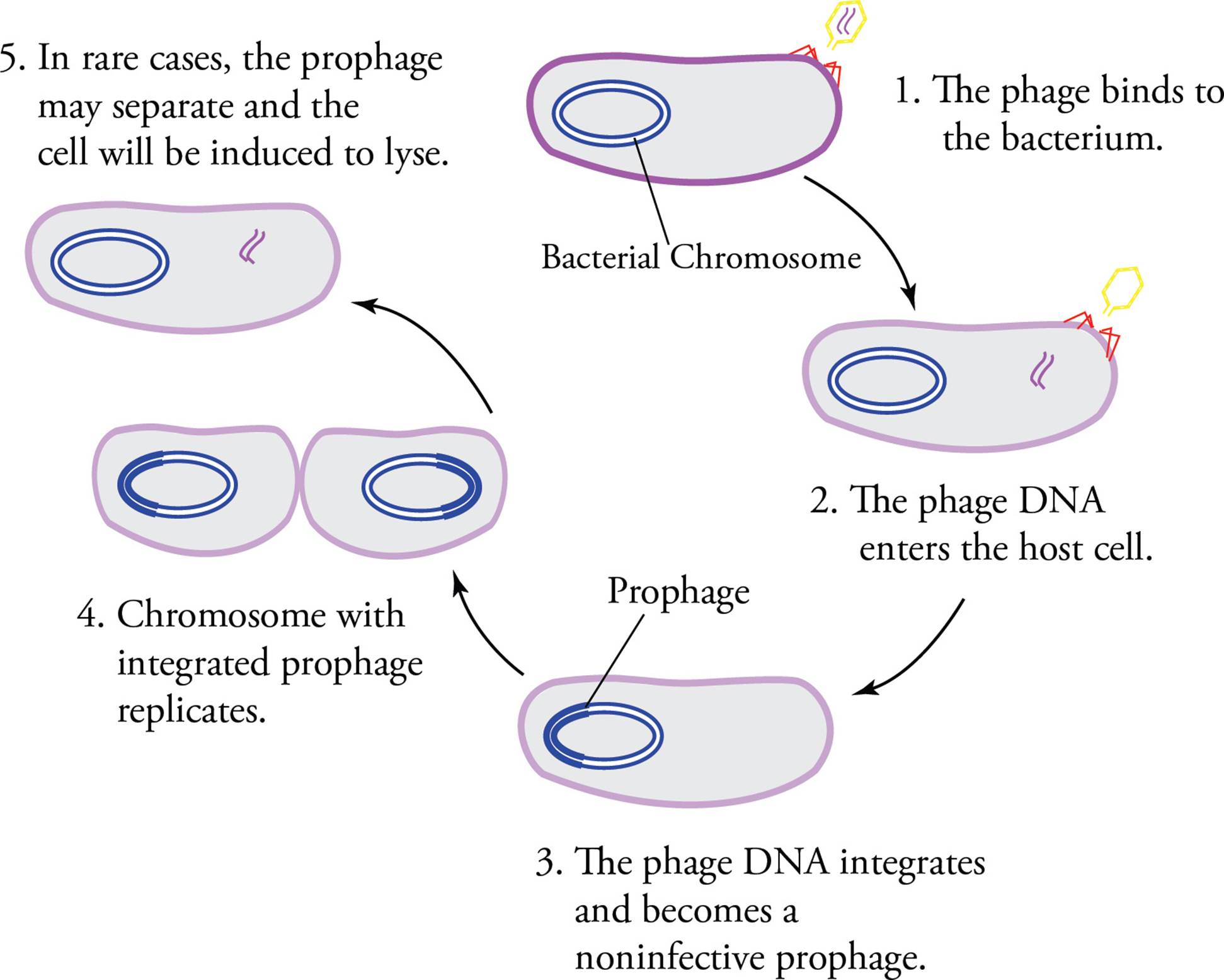
Figure 5 The Lysogenic Cycle
Replication of Animal Viruses
There are a number of differences between phages and viruses which infect animal cells. (Animal viruses don’t have a special name like “phage.”) The general outline of the viral life cycle, however, remains the same. The virus must specifically bind to a proper host cell, release its genetic material into the host, take over host machinery, replicate its genome, synthesize capsid components, assemble itself, and finally escape to infect a new cell.
Animal cells have proteins on the surface of their plasma membranes that serve as specific receptors for viruses. These receptors play a role in normal cellular function; they do not exist simply for the benefit of the virus. Part of the tissue-specificity of animal viruses is due to the distribution of receptors necessary for adsorption. For example, the binding of the HIV virus protein gp120 to a T cell membrane protein termed CD4 is one of the first steps in HIV infection.
• Would treatment of an HIV-infected person with a soluble form of CD4 protein affect the infectivity of the virus?17
• Mutation of the cell-surface receptor that viruses attach to would be a means for an organism to become resistant to viral infection. Why is this mechanism not common?18
• Treatment of an enveloped animal virus with a mild detergent solubilizes several proteins from the virus, although the genome does not become accessible. Which one of the following is consistent with this scenario?19
A) Some of the proteins that are released by detergent may be encoded by the genome of the infected cell.
B) The infectivity of the virus is not affected by detergent treatment.
C) The proteins released by detergent are capsid proteins.
D) All the proteins released by the detergent are encoded by the viral genome.
The next step in the infection of an animal cell is penetration into the cell, just as in bacterial infection by a phage. Many animal viruses enter cells by endocytosis (a process whereby the host cell engulfs the virus and internalizes it). [Why don’t phages enter their hosts by endocytosis?20] Once inside the host, the viral genome is uncoated, meaning it is released from the capsid. Alternatively, some viruses fuse with the plasma membrane to release virus into the cytoplasm. From this point, an animal virus may enter either a lytic cycle, a lytic-like cycle called the productive cycle, or a lysogenic cycle.
The lytic cycle in animal viruses is the same as in phages. The productive cycle is similar to the lytic cycle but does not destroy the host cell. It is possible because enveloped viruses exit the host cell by budding through the host’s cell membrane, becoming coated with this membrane in the process. Budding does not necessarily destroy a cell since the lipid bilayer membrane can reseal as the virus leaves. Finally, in the animal virus lysogenic cycle the dormant form of the viral genome is called a provirus (analogous to a prophage). For example, Herpes simplex I is the virus that causes oral herpes. After infection, it may remain dormant as a provirus for an indefinite period of time. Then one day, usually when the host encounters stress (e.g., lack of sleep, upcoming professional school entrance exams, etc.), the virus reactivates.
Viral Genomes
Many factors determine the uniqueness of each virus. The type of genome, possession or lack of an envelope, nature of cell-surface proteins, and type of life cycle are examples. All of these parameters are used in the classification of viruses, and all are potential targets for therapeutic intervention. The nature of the genome is perhaps the most important of these and has important consequences for how infection by each virus proceeds. In the following discussion we will look at a few viral genomes with an eye to what proteins the virus must encode or actually carry in its capsid based on its genome type. Our purpose is not to provide new information, but rather to demonstrate what conclusions can be drawn from what you already know (typical MCAT passage material). Do not memorize, but rather read for comprehension. We will not discuss ds-RNA or ss-DNA genomes, but by the end of this section you should be able to imagine components they might require.
(+) RNA Viruses
—must encode RNA-dependent RNA pol (and do not have to carry it).
A (+) RNA virus, with a single-stranded RNA genome, is the simplest imaginable type of viral genome. (A piece of single-stranded viral RNA which serves as mRNA is called (+) RNA.) As soon as the (+) RNA genome is in the host cell, host ribosomes begin to translate it, creating viral proteins. The viral genome acts directly as mRNA. The technical way to describe this scenario is to say the genome is infective, meaning injecting an isolated genome into the host cell will result in virus production. In order for the virus to replicate itself, one of the proteins it encodes must be an RNA-dependent RNA polymerase, the role of which is __?21 (+) RNA viruses cause the common cold, polio, and rubella. [Will infectious virus be produced if the genome of an enveloped (+) strand RNA virus is added to an extract prepared from the cytoplasm of eukaryotic cells that retains translational activity but lacks DNA replication or transcription of host genes?22 If a viral genome is (+) strand RNA, what is used as a template by the RNA-dependent RNA polymerase?23]
(–) RNA Viruses
—must carry RNA-dependent RNA pol (and, of course, encode it too).
The genome of a (–) RNA virus is complementary to the piece of RNA that encodes viral proteins. In other words, the genome of a (–) RNA virus is the template for viral mRNA production. If host ribosomes translate (–) RNA, useless polypeptides will be made. Hence, the virus must not only encode an RNA-dependent RNA polymerase, it must actually carry one with it in the capsid. When the virus enters the host cell, this enzyme will create a (+) strand from the (–) genome. Then the viral life cycle can proceed. (–) RNA viruses cause rabies, measles, mumps, and influenza. [Do (–) strand RNA viruses use host enzymes to catalyze RNA production in transcription or in replication of the genome?24]
Retroviruses
—must encode reverse transcriptase.
HIV, the virus that causes AIDS, and HTLV (Human T-cell Leukemia Virus) are examples of retroviruses. These are (+) RNA viruses which undergo lysogeny. In other words, they integrate into the host genome as proviruses. In order to integrate into our double-stranded DNA genome, a viral genome must also be composed of double-stranded DNA. Since these viral genomes enter the cell in an RNA form, they must undergo reverse transcription to make DNA from an RNA template. This snubbing of the central dogma is accomplished by an RNA-dependent DNA polymerase (“reverse transcriptase”) encoded by the viral genome. Retroviruses are theoretically not required to carry this enzyme, only to encode it. [Why?25] The three main retroviral genes are gag (codes for viral capsid proteins), pol (polymerase codes for reverse transcriptase) and env (envelope codes for viral envelope proteins). [After integration of a retrovirus into the cellular genome, a reverse transcriptase inhibitor is added to the cell. Will the production of new viruses be blocked?26]
Double-stranded DNA Viruses
—often encode enzymes required for dNTP synthesis and DNA replication.
These viruses often have large genomes that include genes for enzymes involved in deoxyribonucleotide synthesis (which we do whenever we make DNA) and DNA replication. [Given the limited information that viruses may contain in their genomes, why carry around genes for an enzyme possessed by the host?27 Why don’t RNA viruses do this?28 What is a factor likely to limit the size of RNA genomes?29 Some DNA viruses induce infected host cells to enter mitosis and may even override cellular inhibition of cell division so strongly that the cell becomes cancerous; what is the advantage to the virus of inducing host-cell division?30]
• Adenoviruses have a single linear ds-DNA genome, which contains a number of different promoters that are regulated during infection. Although transcription is carried out by cellular RNA polymerase, the viral E1A gene product is required for transcription of most viral genes. If the E1A gene is deleted from the virus or if the gene product is inactivated, viral infection is unable to proceed. Adenoviruses also encode much of their own replication machinery, including DNA polymerase. If two different adenoviruses infect the same cell, one with a deleted E1A gene and another with a deleted DNA polymerase gene, will successful infection of the cell result?31
6.2 SUBVIRAL PARTICLES
Some infectious agents are even smaller and simpler than viruses and are termed subviral particles. These include prions and viroids.
Prions
As infectious agents, prions do not strictly follow the Central Dogma because they are self-replicating proteins [Why does this violate the Central Dogma?32]. The prion itself is a misfolded version of a protein that already exists (see Figure 6).
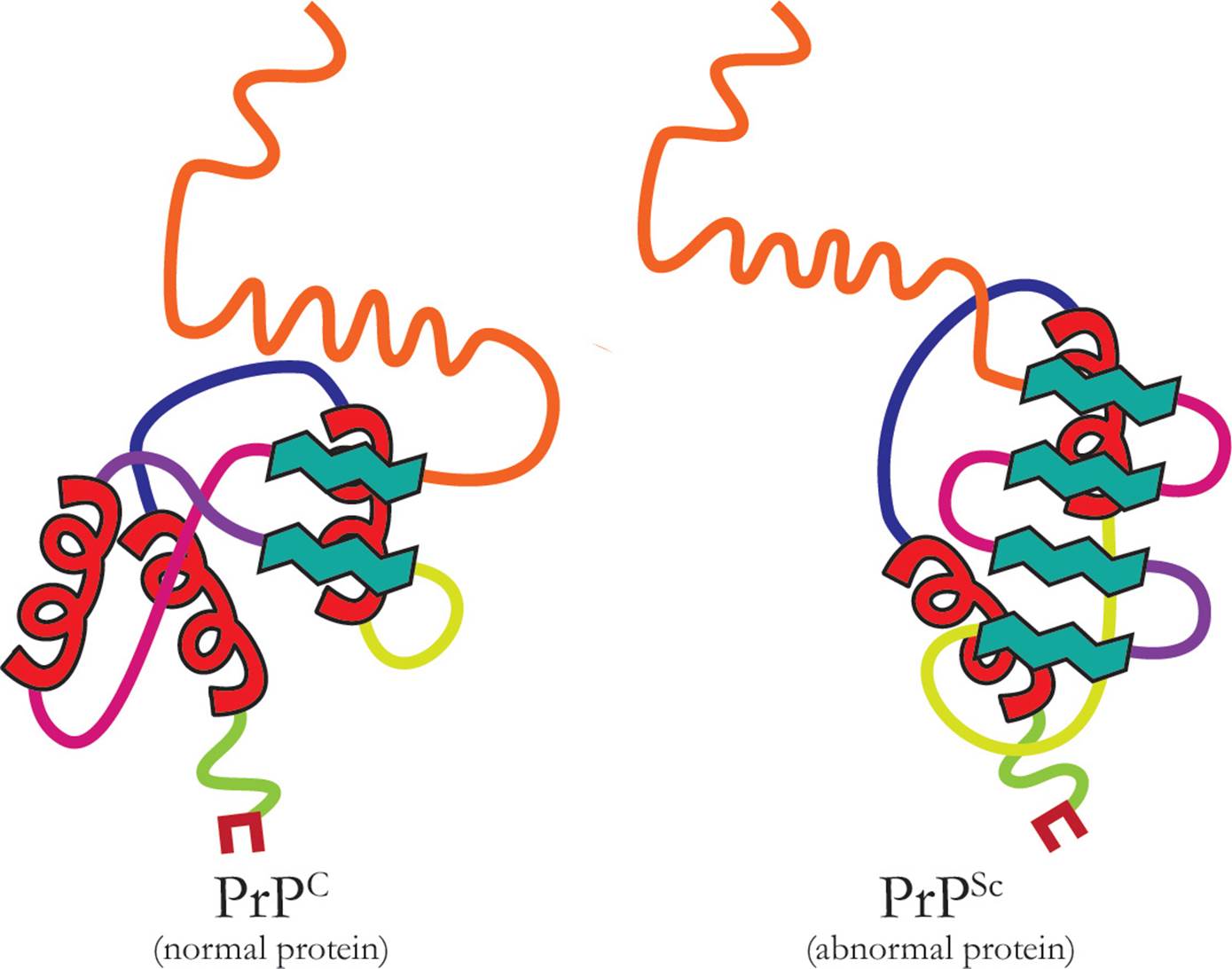
Figure 6 Comparison of the PrPC structure to the PrPSc structure
When the normally folded protein (designated PrPC) comes into contact with the prion (designated PrPSc), the prion acts as a template; the shape of the normal protein is altered and it too becomes infectious. Prions are responsible for a class of diseases in mammals referred to as the transmissible spongiform encephalopathies (TSEs). These diseases cause degeneration in the nervous system, especially the brain where characteristic holes develop, and are always fatal. The misfolded proteins are found in the nervous tissues and are very resistant to degradation by chemicals or heat, making them hard to destroy. Bovine spongiform encephalopathy (BSE, commonly called mad cow disease) is the prion disease found in cows; this was originally transmitted to cows from sheep because all types of tissue from sheep, including the brain, is used as a supplement in the feed for other farm animals. Though much less common, the disease kuru follows a similar transmission path in humans; it is only found in a limited number of tribes where consumption of the body, particularly of the brain, is part of honoring the dead (since identification of the transmission route, this practice has stopped and kuru has virtually disappeared).
However, prion diseases can also be genetically linked, through mutations in the gene that codes for the prion protein. For example, fatal familial insomnia (FFI) is an autosomal dominant condition inherited on chromosome 20, and Creutzfeldt Jakob disease (CJD) is also inherited. It is also possible for these diseases to arise spontaneously (through mutation) in someone with no prior family history. In general, however, prion diseases are very rare, striking only 1–2 people per million.
Whether transmitted, inherited or spontaneously arising, prion diseases are characterized by their very long incubation periods, which can be several months to years in animals and several years to decades in humans. The misfolded proteins cause the destruction of neurons, particularly in the central nervous system, leading to loss of coordination, dementia, and death. Diagnosis is difficult, in part because of the long incubation periods and in part because the symptoms can be indicative of other conditions.
Viroids
Viroids consist of a short piece of circular, single-stranded RNA (200-400 bases long) with extensive self-complementarity (i.e., it can base-pair with itself to create some regions that are double-stranded, see Figure 7). Generally they do not code for proteins and they lack capsids. Some viroids are catalytic ribozymes, while others, when replicated, produce siRNAs that can silence normal gene expression.
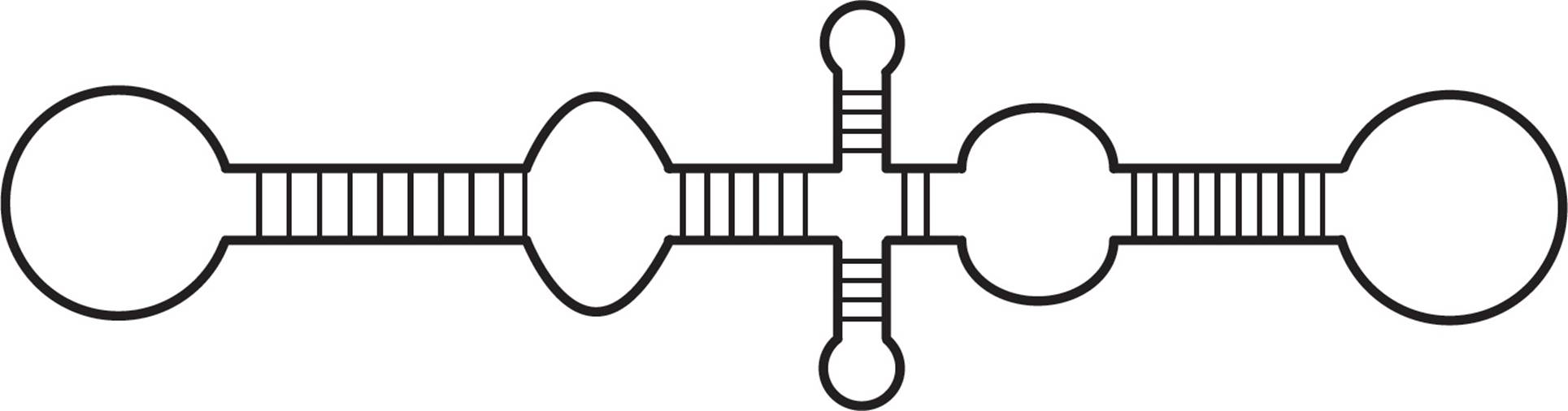
Figure 7 Structure of a viroid showing double-stranded regions
Replication of some viroids shares similarity to the replication of RNA viruses. A viroid RNA-dependent RNA polymerase synthesizes a (–) strand which is circularized by an RNA ligase derived from the host; this is then used as the round, rolling template to make more (+) copies that match the original RNA viroid sequence. An alternative to this mechanism leaves the (–) stand in a more linear state where it can still act as a template for (+) strand creation, and then become circularized. In other cases, viroids somehow hijack the cell’s DNA dependent RNA polymerase and direct it to read RNA templates. This mechanism is not well understood.
Most of the diseases caused by viroids are found in plants. The only human disease linked to viroids is Hepatitis D. The Hepatitis D viroid can only enter hepatocytes (liver cells) if it is contained in a capsid with a binding protein; since viroids do not have capsids, successful Hepatitis D infection required coinfection with Hepatitis B, from which it derives its capsid.
6.3 PROKARYOTES (DOMAIN BACTERIA)
Cell Theory
Advances in microbiology have been made possible by advancing technologies in magnification. Once humans were able to utilize basic, if crude, microscopy, the cell as the monomer of tissues and organs could be studied. In 1655, this led the English scientist, Robert Hooke, to define the Cell Theory based on his studies of cork. Its tenets are as follows:
1) All living organisms are composed of one or more cells and their products.
2) Cells are the monomer for any organism.
3) New cells arise from pre-existing, living cells.
Though these basic principles are still true, more modern extensions of Cell Theory also include the idea that no matter what the species, the chemical composition of cells is similar, that DNA is the source of hereditary programming information passed from cell to cell, that an organism’s activity is determined by the total activity of its cells, and that biochemical energy flow occurs within cells. These additional principles have been explored and verified due to vast improvements both in microscopy as well as biochemical and genetic testing.
All living organisms (which does not include viruses) can be classified as either prokaryotes or eukaryotes. The classification of organisms into these groups is based on examination of their internal cellular structure. Representatives from both groups are able to carry out the basic biochemical processes of photosynthesis, the Krebs cycle, and oxidative phosphorylation to produce ATP. The primary feature of prokaryotes that distinguishes them from eukaryotes is that they do not contain membrane-bound organelles (nucleus, mitochondria, lysosomes, etc.). Prokaryote means “before the nucleus,” and the lack of a nucleus indicates that prokaryotes are evolutionarily the oldest domains. Unlike viruses however, prokaryotes possess all of the machinery required for life. They are true cells; true living organisms. The prokaryotes include bacteria, archea (extremophiles), and blue-green algae(cyanobacteria).
The classification of living organisms, taxonomy, is an important part of biology because it is used to determine the evolutionary relationship of organisms to one another. The largest taxonomic division is the domain. There are three recognized domains: Bacteria, Archea, and Eukarya. Domains Bacteria and Archea include prokaryotic organisms, and Domain Eukarya includes eukaryotic organisms. Each domain can be further subdivided into kingdoms. Currently there are three well-recognized eukaryotic kingdoms (Animalia, Plantae, and Fungi), and great debate over the number of kingdoms that should be present in the other prokaryotic domains and in the single-celled eukaryotes (protists).
In this section, we will begin to study the most basic and ancient of organisms, the prokaryotes.
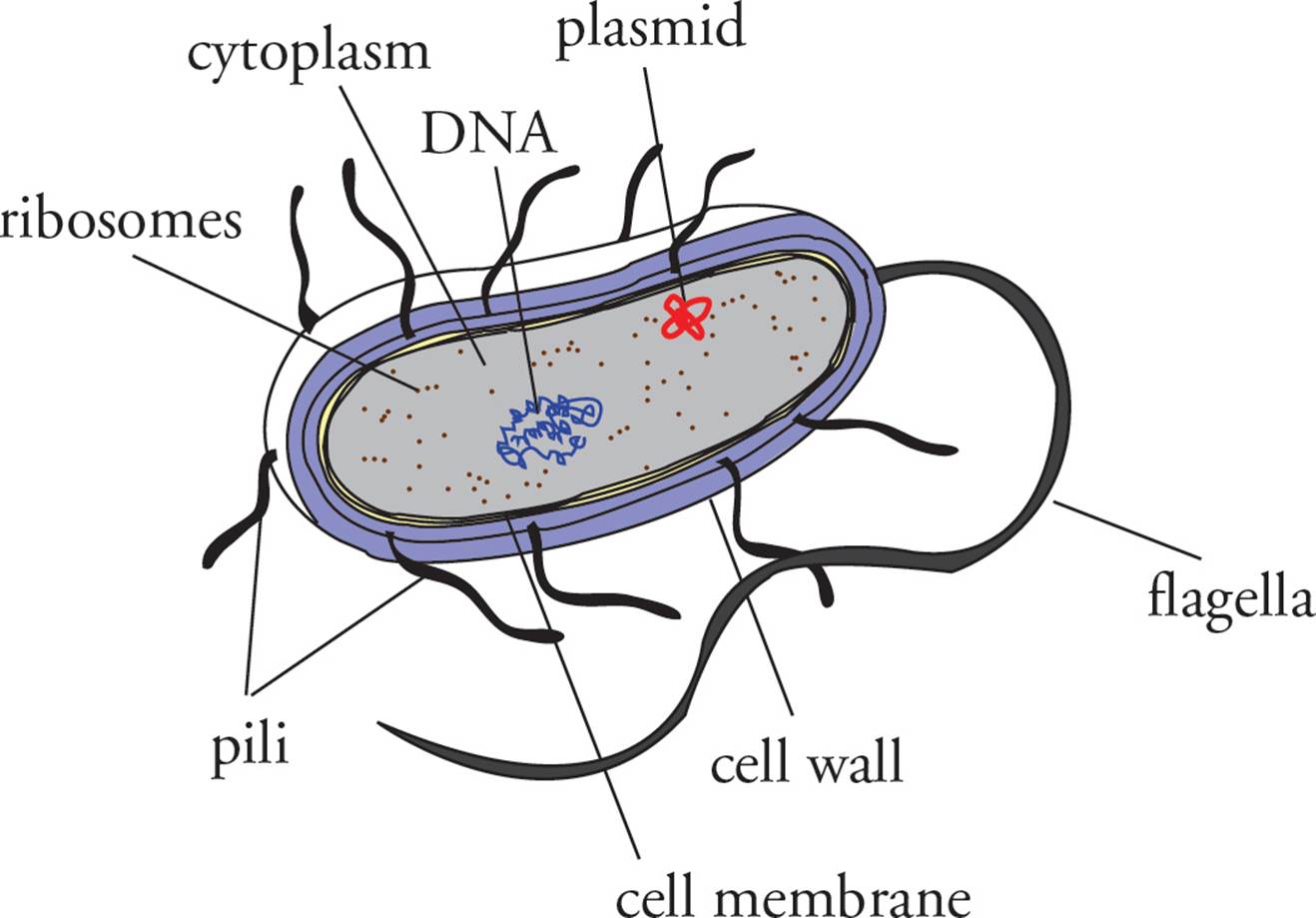
Figure 8 A Prokaryote
Bacterial Structure and Classification
Contents of the Cytoplasm
In this section we will tour the bacterial cell from the inside out. Unlike a eukaryotic cell, there are no membrane-bound organelles in prokaryotic cells (note that ribosomes, which are not membrane-bound, are found in bacteria). The prokaryotic genome is a single double-stranded circular DNA chromosome.33 It is not located in a nucleus and is not associated with histone proteins, as the eukaryotic genome is. In bacteria, transcription and translation occur in the same place, at the same time. Ribosomes begin to translate mRNA before it is completely transcribed. Many ribosomes translating a single piece of mRNA form a structure known as a polyribosome.
[In Figure 9 below, is the free end of the mRNA the 3′ or the 5′ end? Which end of the nascent polypeptides is the free end?34] Remember that the bacterial ribosome is structurally different from the eukaryotic ribosome, though both function the same way. The differences allow us to prescribe various antibiotics which interfere with bacterial translation without disrupting our own. (Examples are streptomycin and tetracycline, which only bind to bacterial ribosomes.)
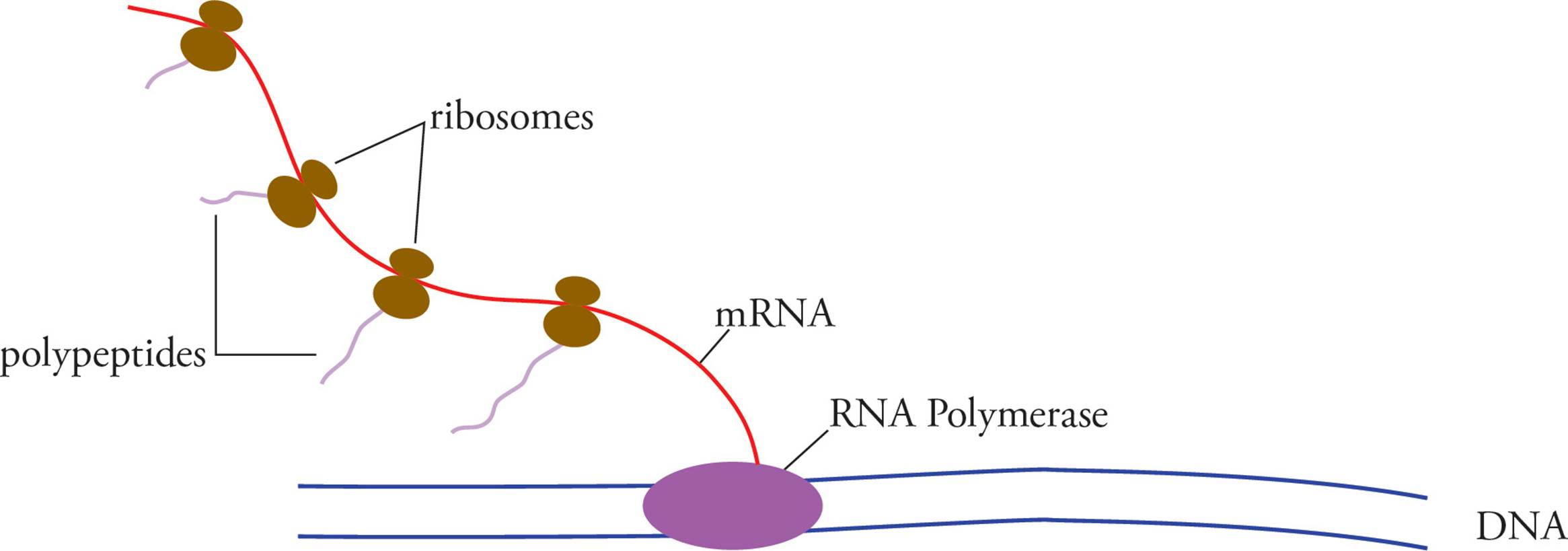
Figure 9 A Prokarytic Polyribosome
One last genetic element that can be found in prokaryotic cells is the plasmid. This is a circular piece of double-stranded DNA which is much smaller than the genome. Plasmids are referred to as extrachromosomal genetic elements. They often encode gene products which may confer an advantage upon a bacterium carrying the plasmid. For example, plasmids frequently carry antibiotic-resistance genes (genes that encode proteins which can break down antibiotics). Many plasmids are capable of autonomous replication, which means that a single plasmid molecule within a bacterial cell may cause itself to be replicated into many copies. Plasmids are important not only because they may encode advantageous gene products, but also because they orchestrate bacterial exchange of genetic information, or conjugation, which is discussed below.
Bacterial Shape
Bacteria are often classified according to their shape. The three shapes and their proper names are organized in the following table:

Table 1 Bacterial Classification by Shape
The Cell Membrane and the Cell Wall
The bacterial cytoplasm is bounded by a lipid bilayer which is similar to our own plasma membrane. Outside the lipid bilayer is a rigid cell wall. It provides support for the cell, preventing lysis due to osmotic pressure. (As we will discuss in Chapter 7, animal cells lack a cell wall. They deal with the problem of osmotic pressure by continuously pumping ions across the cell membrane.) The bacterial cell wall is composed of peptidoglycan, a complex polymer unique to prokaryotes. It contains cross-linked chains made of sugars and amino acids, including D-alanine, which is not found in animal cells (our amino acids have the L configuration). The bacterial cell wall is the target of many antibiotics, such as penicillin. The enzyme lysozyme, which is found in tears and saliva and made by lytic viruses, destroys the peptidoglycan in the bacterial cell wall, resulting in an osmotically fragile structure called a protoplast. [Would a protoplast moved from salt water to fresh water shrivel or burst?35]
Gram Staining of the Cell Wall
As part of our tour of the bacterial cell, we will say a word about classification of bacteria according to two different types of cell wall. The method of classification is derived from the extent to which bacteria turn color in a procedure termed Gram staining. The two groupings are Gram-positive, which stain strongly (a dark purple color) and Gram-negative bacteria, which stain weakly (a light pink color).
Gram-positive bacteria have a thick peptidoglycan layer outside of the cell membrane and no other layer beyond this. Gram-negative bacteria have a thinner layer of peptidoglycan in the cell wall but have an additional outer layer containing lipopolysaccharide. The intermediate space in Gram-negative bacteria between the cell membrane and the outer layer is termed the periplasmic space, in which are sometimes found enzymes that degrade antibiotics (see Figure 10). The increased protection of Gram-negative bacteria from the environment is reflected in their weak staining, as well as in their increased resistance to antibiotics. [Which bacteria would be more susceptible to lysis when treated with lysozyme: Gram-positive or Gram-negative?36]
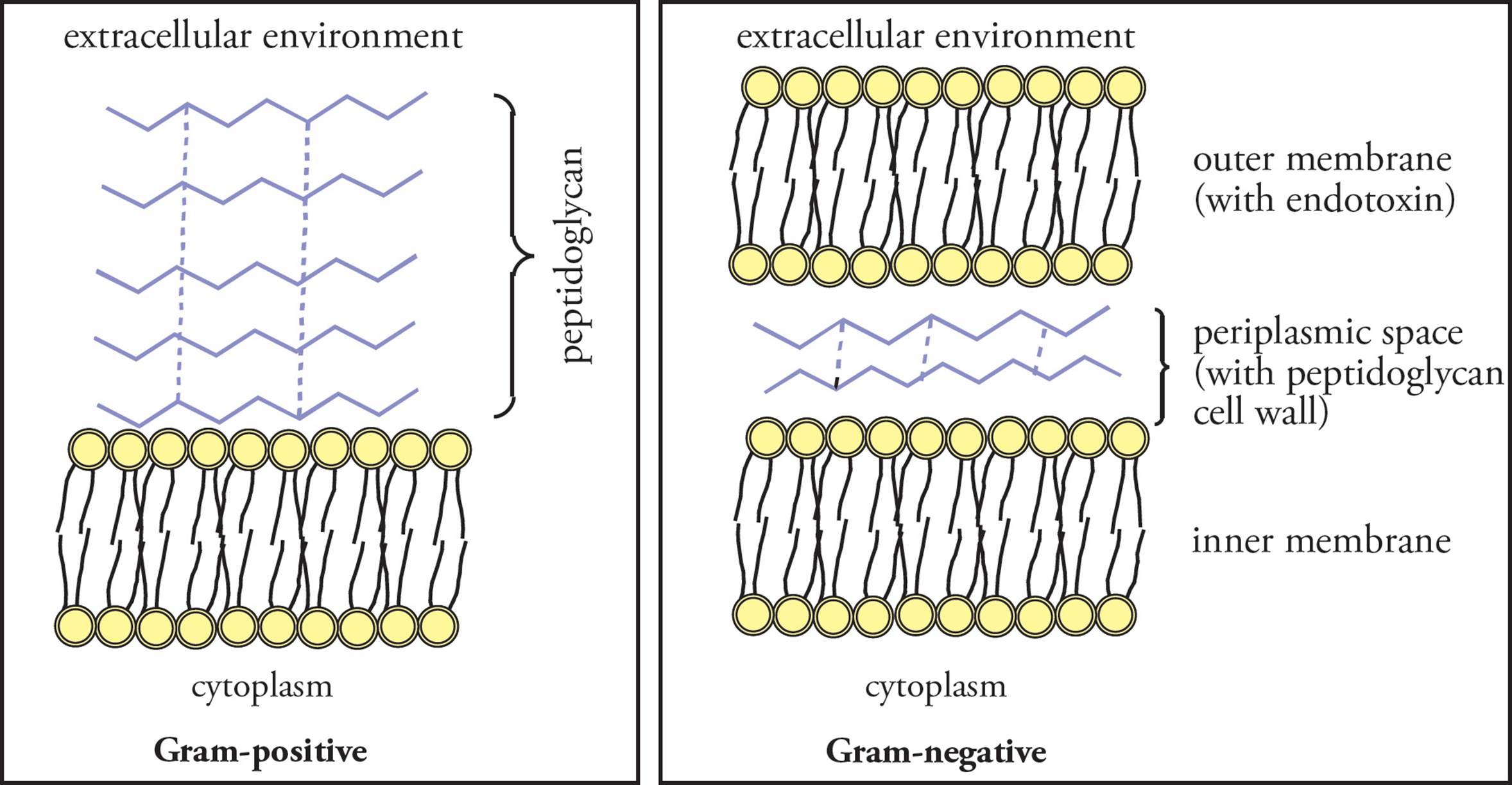
Figure 10 Gram-positive vs. Gram-negative Bacteria
Endotoxins vs. Exotoxins
Endotoxins are normal components of the outer membrane of Gram-negative bacteria that aren’t inherently poisonous. However, they cause our immune system to have such an extreme reaction that we may die as a result. Endotoxins cause the most trouble when many bacteria die and their disintegrated outer membranes are released into the circulation. When this occurs, cells of the immune system release so many chemicals that the patient goes into what is called septic shock, in which much of the aqueous portion of the blood is leaked into the tissues causing a drop in blood pressure, and other problems, which may be fatal. Endotoxins can have various chemical structures including lipopolysaccharide, which contains sugars bound to lipids.
Exotoxins are very toxic substances secreted by both Gram-negative and Gram-positive bacteria into the surrounding medium. Exotoxins help the bacterium compete with other bacterial species, such as normal inhabitants of the mammalian gut. Some diseases that are caused by exotoxins are botulism, diptheria, tetanus, and toxic shock syndrome.
The Capsule
Another attribute which only some bacteria have is the capsule or glycocalyx. This is a sticky layer of polysaccharide “goo” surrounding the bacterial cell and often surrounding an entire colony of bacteria. It makes bacteria more difficult for immune system cells to eradicate. It also enables bacteria to adhere to smooth surfaces such as rocks in a stream or the lining of the human respiratory tract.
Flagella
Another item only some bacteria have are long, whip-like filaments known as flagella, which are involved in bacterial motility. [Can viruses move via flagellar propulsion to find host cells?37] A bacterium which possesses one or more flagella is said to be motile, because flagella are the only means of bacterial locomotion. Bacteria may be monotrichous (meaning they have a flagellum located at only one end), amphitrichous (meaning they have a flagellum located at both ends), or peritrichous (meaning that they have multiple flagella). The following is which?38

The structure of the flagellum is fairly complicated, with components encoded by over 35 genes, but it can be broken down into a few major components: the filament, the hook, and the basal structure (Figure 11). The basal structure contains a number of rings that anchor the flagellum to the inner and outer membrane (for a Gram-negative bacterium) and serve to rotate the rod and the rest of the attached flagellum in either a clockwise or counterclockwise manner. The most important thing to remember about the prokaryotic flagellum is that its structure is different from the eukaryotic one (which contains a “9 + 2” arrangement of microtubules, discussed in Chapter 7).
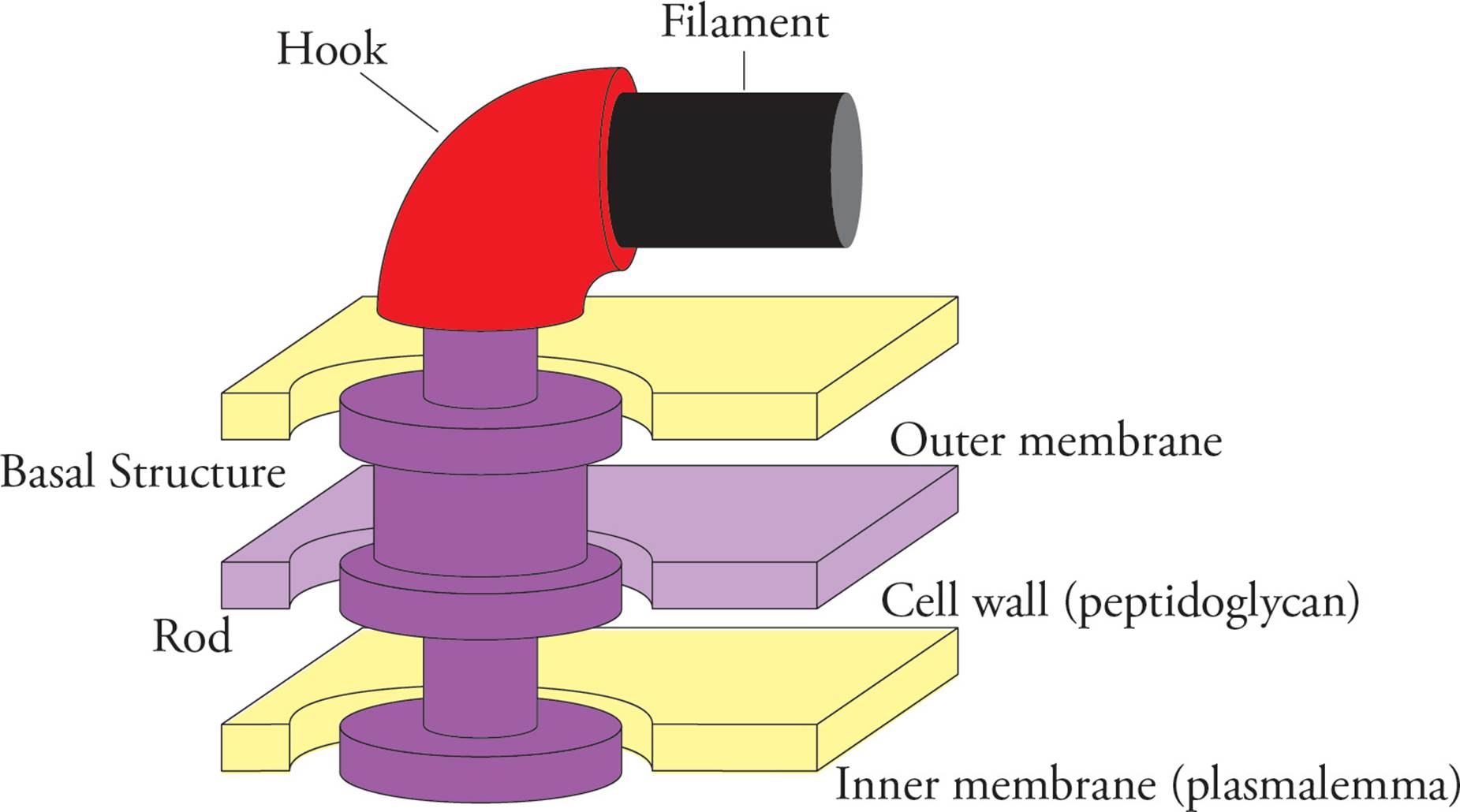
Figure 11 The Prokaryotic Flagellum
The rotation of the rod requires a large amount of energy (that is, ATP), which is supplied by the diffusion of H+ down the proton gradient generated across the inner membrane by electron transport. Bacterial motion can be directed toward attractants, such as food, or away from toxins, such as acid, in a process termed chemotaxis. The connection between chemotaxis and flagellar propulsion is dependent upon chemoreceptors on the cell surface that bind attractants or repellents and transmit a signal which influences the direction of flagellar rotation. A good analogy would be the blind man’s bluff game played by children, in which a person is blindfolded and moves randomly but selects among favorable or unfavorable movements toward the goal based on the responses “warmer” or “colder” (like chemoreceptors binding attractant or repellent and sending a signal to the bacteria to tumble or not to tumble). The response of flagellar rotation to chemical attractants (or repellents) is not dependent on an absolute concentration, but to a change in the concentration over time. Thus, as the bacterium moves through the solution it is able to detect whether it is moving toward or away from the highest concentration and respond accordingly.
Pili
Pili are long projections on the bacterial surface involved in attaching to different surfaces. The sex pilus is a special pilus attaching F+ (male) and F– (female) bacteria which facilitates the formation of conjugation bridges(discussed below). Fimbriae are smaller structures that are not involved in locomotion or conjugation but are involved in adhering to surfaces. [What other bacterial structure is involved in adhering to surfaces? Is it possible that the fimbriae play a role in infection by pathogenic organisms?39]
Bacterial Growth Requirements and Classification
Temperature
Another characteristic of bacteria used to categorize them is their ability to tolerate environmental variables, such as temperature. Though bacteria as a group can grow at a wide range of temperatures, each species has an optimal growth temperature. If the temperature is too high or too low, bacteria fail to grow and may be killed, hence the use of boiling to kill bacteria and refrigeration to slow bacterial growth and prevent food spoilage. Most bacteria favor mild temperatures similar to the ones that humans and other organisms favor (30°C); they are called mesophiles (moderate temperature lovers). Thermophiles (heat lovers) can survive at temperatures up to 100°C in boiling hot springs or near geothermal vents in the ocean floor. Bacteria that thrive at very low temperatures (near 0°C) are termed psychrophiles (cold lovers). [How might a decrease in temperature increase the bacterial growth rate?40]
Nutrition
Bacteria can be classified according to their carbon source and their energy source. “Troph” is a Latin root meaning “eat.” Autotrophs utilize CO2 as their carbon source. Heterotrophs rely on organic nutrients (glucose, for example) created by other organisms. Chemotrophs get their energy from chemicals. Phototrophs get their energy from light; not only plants but also some bacteria do this. Each bacterium is either a chemotroph or a phototroph and is either an autotroph or a heterotroph. There are thus four types of bacteria:
1) Chemoautotrophs build organic macromolecules from CO2 using the energy of chemicals. They obtain energy by oxidizing inorganic molecules like H2S.
2) Chemoheterotrophs require organic molecules such as glucose made by other organisms as their carbon source and for energy. (We are chemoheterotrophs.)
3) Photoautotrophs use only CO2 as a carbon source and obtain their energy from the Sun. (Plants are photoautotrophs.)
4) Photoheterotrophs are odd in that the get their energy from the Sun, like plants, but require an organic molecule made by another organism as their carbon source.
• A bacterium that causes an infection in the bloodstream of humans is most likely to be classified as which one of the following?41
A) Chemoautotroph
B) Photoautotroph
C) Chemoheterotroph
D) Photoheterotroph
• Which one of the following categories best describes an organism which uses sunlight to drive ATP production but cannot incorporate carbon dioxide into sugars?42
A) Chemoautotroph
B) Photoautotroph
C) Chemoheterotroph
D) Photoheterotroph
Growth Media
The environment in which bacteria grow is the medium (plural: media). In the lab, the most common solid medium is agar, a firm transparent gel made from seaweed. Bacteria live in the agar but do not metabolize it. The agar is usually kept in a clear plastic plate called a Petri dish, and the process of putting bacteria on such a plate is called plating. When one bacterium is plated onto a dish, if it grows, it will eventually give rise to many progeny in an isolated spot called a colony. Minimal medium contains nothing but glucose (in addition to the agar). More key terms: A wild-type bacterium (or a wild-type strain) is one which possesses all the characteristics normal to that particular species. The dense growth of bacteria seen in laboratory Petri dishes is known as a bacterial lawn. A plaque is a clear area in the lawn. Plaques result from death of bacteria and are caused by lytic viruses or toxins.
Bacteria can reproduce very rapidly, provided that the conditions of their environment are favorable and nutrients are abundant. The doubling time is the amount of time required for a population of bacteria to double its number. It ranges from a minimum of 20 minutes for E. coli to a day or more for slow growers, such as the bacteria responsible for tuberculosis and leprosy. The doubling time of a bacterial species will vary, depending upon the availability of nutrients and other environmental factors.
One other important term in bacterial nutrition is auxotroph (don’t confuse this term with autotroph). This is a bacterium which cannot survive on minimal medium because it can’t synthesize a molecule it needs to live. Hence, it requires an auxiliary trophic substance to live. For instance, a bacterium which is auxotrophic for arginine won’t form a colony when plated onto minimal medium, but if the medium is supplemented with arginine, a colony will form. This arginine auxotrophy is denoted arg–. Auxotrophy results from a mutation in a gene coding for an enzyme in a synthetic pathway.
Bacteria can be differentiated not only by what substances they require, but also by what substances they are capable of metabolizing for energy. For instance, a strain of bacteria may be capable of surviving on minimal medium that has the disaccharide lactose as the only carbon source (no glucose). This would be denoted lac+. Mutation in a gene for the enzyme lactase would impair the bacterium’s ability to survive on lactose-only medium. A bacterial strain incapable of growing with lactose as its only carbon source would be denoted lac–. Genetic exchange between bacteria by means of conjugation, transduction, or transformation (discussed below) can remedy these disabilities.
Oxygen Utilization and Tolerance
Oxygen metabolism is aerobic metabolism. Bacteria which require oxygen are called obligate aerobes. Bacteria which do not require oxygen are called anaerobes. There are three subcategories: facultative anaerobes will use oxygen when it’s around, but don’t need it. [How much more ATP can they make per glucose molecule when O2 is present?43] Tolerant anaerobes can grow in the presence or absence of oxygen but do not use it in their metabolism. Obligate anaerobes are poisoned by oxygen. This is because they lack certain enzymes necessary for the detoxification of free radicals which form spontaneously whenever oxygen is around.44 Obligate anaerobes commonly infect wounds.
• If a bacterium cannot use oxygen as an electron acceptor, is it an obligate anaerobe, a tolerant anaerobe, a facultative anaerobe, or is it not possible to distinguish based on the information given?45
• A sample of bacteria is evenly mixed into a cool liquid agar nutrient mix in the absence of oxygen and then poured into a glass-walled tube that is open to the atmosphere on top. When the agar mix cools, it solidifies, and bacterial growth is observed as shown below. How would you classify the bacteria in terms of oxygen utilization and tolerance? (Note: Agar is practically impermeable to oxygen.)46
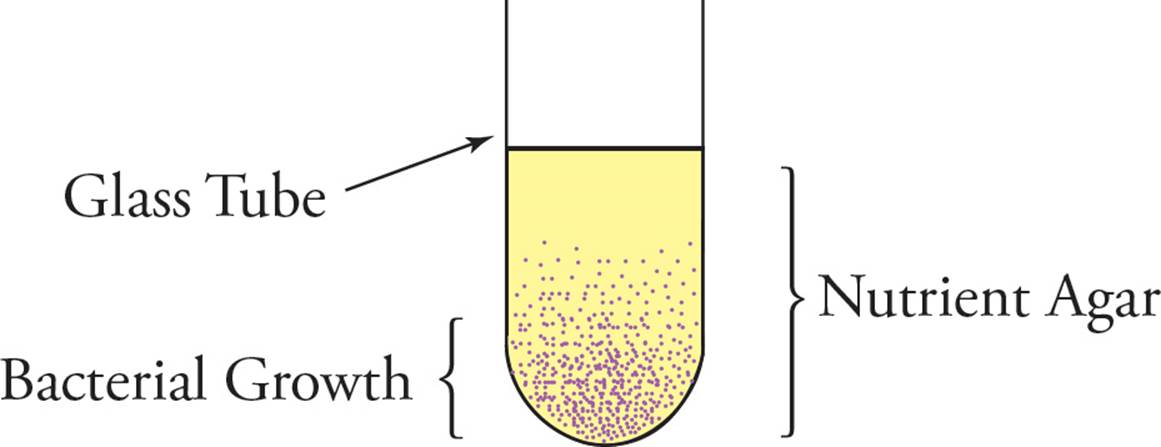
Fermentation vs. Respiration
This was covered in Chapter 4. To briefly review, respiration is glucose catabolism with use of an inorganic electron acceptor such as oxygen. In contrast, fermentation is glucose catabolism which does not use an electron acceptor such as O2; instead, a reduced by-product of glucose catabolism such as lactate or ethanol is given off as waste. [Why is fermentation necessary whenever an external electron acceptor is not used?47]
Anaerobic Respiration
This is not a contradiction in terms! It refers to glucose metabolism with electron transport and oxidative phosphorylation relying on an external electron acceptor other than O2. For example, instead of reducing O2 to H2O, some anaerobic bacteria reduce SO42– to H2S, or CO2 to CH4. Nitrate (NO3–) is another possible electron acceptor.
• In an experiment, facultative anaerobic bacteria that are growing on glucose in air are shifted to anaerobic conditions. If they continue to grow at the same rate while producing lactic acid, then the rate of glucose consumption will:48
A) increase 16 fold.
B) decrease 16 fold.
C) decrease 2 fold.
D) not change.
Bacterial Life Cycle
Bacteria reproduce asexually. In asexual reproduction, there is no meiosis, no meiotic generation of haploid gametes, and no fusion of gametes to form a new individual organism. Instead, each bacterium grows in size until it has synthesized enough cellular components for two cells rather than one, replicates its genome, then divides in two. This process in bacteria is also known as binary fission (fission means “to split”). [In prokaryotes, does reproduction increase genetic diversity?49 If a eukaryote reproduces strictly by asexual reproduction, how will this affect the genetic diversity of a population?50 How is asexual reproduction in a eukaryote different from asexual reproduction in a prokaryote?51] Although bacteria do not reproduce sexually, they do possess a mechanism, termed conjugation, for exchanging genetic information (more on this later).
Growth of bacterial populations is described in stages (see Figure 12). Under ideal conditions, bacterial population growth is exponential, meaning that the number of bacterial cells increases exponentially with time. This also means the log of the population size grows linearly with time, hence the name log phase. [If 10 bacteria in log phase are placed in ideal growth conditions and the doubling time is 20 minutes, how many bacteria will there be after four hours?52]
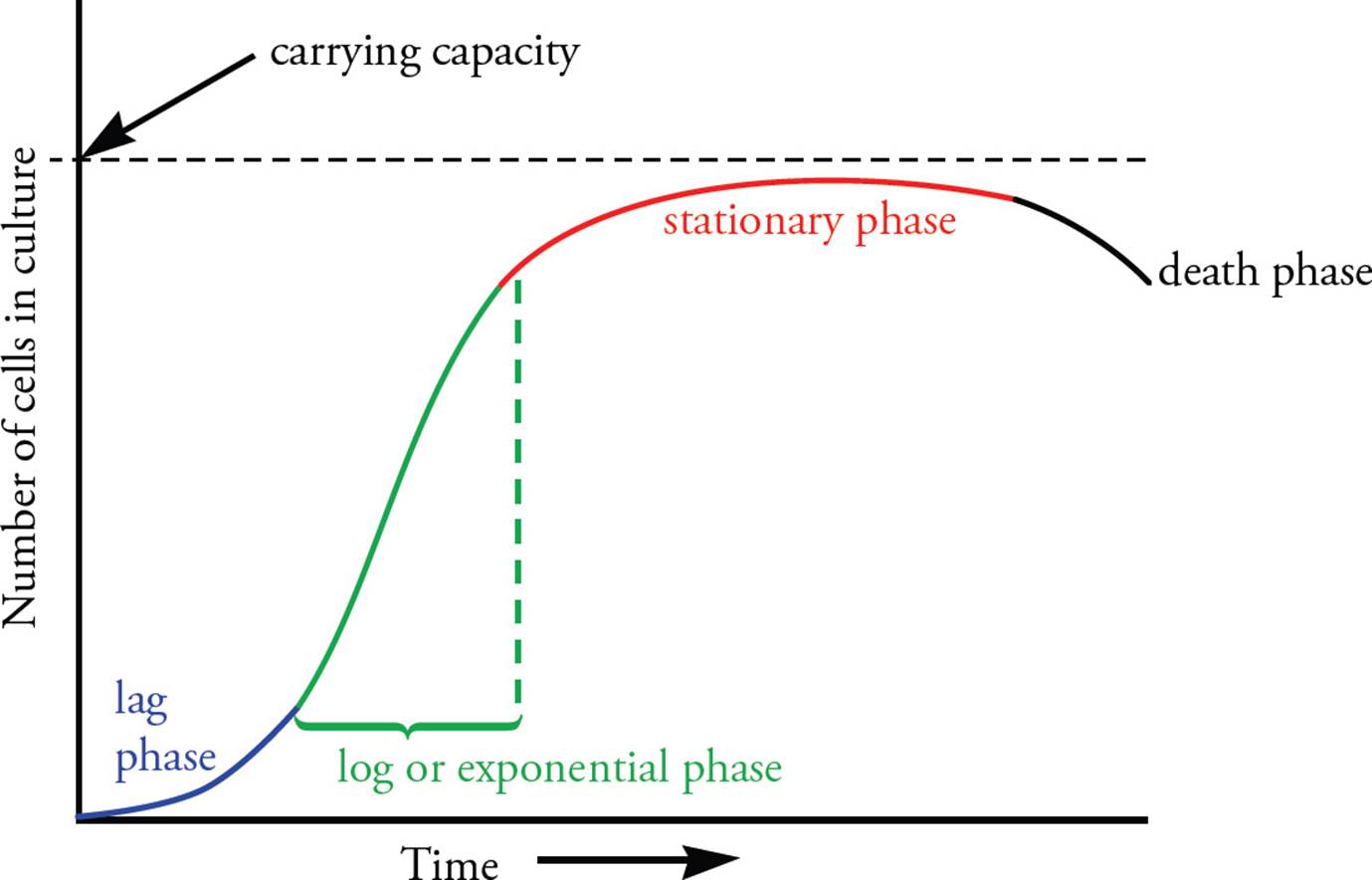
Figure 12 Bacterial Reproduction
Prior to achieving exponential growth, bacteria that were not previously growing undergo a lag phase, during which cell division does not occur even if the growth conditions are ideal.
• If growth conditions are ideal, why wouldn’t cell division occur immediately?53
• Will bacteria that are transferred from a culture that is in log phase to a fresh new culture show a lag phase?54
As metabolites in the growth medium are depleted, and metabolic waste products accumulate, the bacterial population passes from log phase to stationary phase, in which cells cease to divide for lack of nutrients. The maximum population at the stationary phase is referred to as the carrying capacity for that environment. In the last stages of the stationary phase, cell death may occur as a result of the medium’s inability to support growth. [If bacteria are grown in a medium with glucose as the main source of energy, when will the glycolytic pathway be more active: during the lag phase or during the stationary phase?55]
Endospore Formation
Some types of Gram-positive bacteria, such as the bacteria responsible for botulism, form endospores under unfavorable growth conditions. Endospores have tough, thick external shells comprised of peptidoglycan. Within the endospore are found the genome, ribosomes, and RNA which are required for the spore to become metabolically active when conditions become favorable. Endospores are able to survive temperatures above 100°C, which is why autoclaves or pressure cookers are required to completely sterilize liquids and substances that cannot be heated sufficiently in a dry oven. The metabolic reactivation of an endospore is termed germination. A single bacterium is able to form only one spore per cell. Thus, bacteria cannot increase their population through spore formation. [When are bacteria most likely to form endospores: during lag phase, log phase, or stationary phase? Is endospore formation a means for bacteria to reproduce?56]
Genetic Exchange Between Bacteria
Bacteria reproduce asexually, but genetic exchange is evolutionarily favorable because it fosters genetic diversity. Bacteria have three mechanisms of acquiring new genetic material: transduction, transformation, and conjugation. Note that none of these has anything to do with reproduction! Transduction was discussed in Section 6.1 under “lysogenic cycle”; it is the transfer of genomic DNA from one bacterium to another by a lysogenic phage. Transformation refers to a peculiar phenomenon: If pure DNA is added to a bacterial culture, the bacteria internalize the DNA in certain conditions and gain any genetic information in the DNA. Conjugation appears most likely to be related to normal bacterial function, however.
Conjugation
Although bacteria reproduce asexually, they have developed conjugation to exchange genetic information. In conjugation, bacteria make physical contact and form a bridge between the cells. One cell copies DNA, and this copy is transferred through the bridge to the other cell. A key to bacterial conjugation is an extrachromosomal element known as the F (fertility) factor. Bacteria that have the F factor are male, or F+, and will transfer the F factor to female cells. Bacteria that do not contain the F factor are female, F–, and will receive the F factor from male cells to become male. [If all cells in a population are F+, will conjugation occur?57]
The F factor is a single circular DNA molecule. Although much smaller than the bacterial chromosome, the F factor contains several genes, many of which are involved in conjugation itself. [Which cell will produce sex pili: the male cell or the female cell?58] After the male cell produces sex pili and the pili contact a female cell, a conjugation bridge forms. The F factor is replicated and transferred from the F+ to the F– cell. DNA transfer between F+ and F– cells is unidirectional; it occurs in one direction only (see Figure 13).
Although the F factor is an extrachromosomal element, it does sometimes become integrated into the bacterial chromosomes through recombination. A cell with the F factor integrated into its genome is called an Hfr (highfrequency of recombination) cell. [Will an Hfr cell undergo conjugation with an F– cell?59] When an Hfr cell performs conjugation, replication of the F factor DNA occurs as in F+ cells with the extra chromosomal F factor. Since the F factor DNA is integrated in the bacterial genome in Hfr cells, replication of F factor DNA continues into bacterial genes, and these too can be transferred into the F– cell (see Figure 13).
• If bacteria contain only one copy of the bacterial genome, how can recombination occur?60
• If the F factor in an Hfr strain integrates near a gene required for lactose metabolism, is it likely that other genes involved in lactose metabolism will be transferred during conjugation at the same time?61
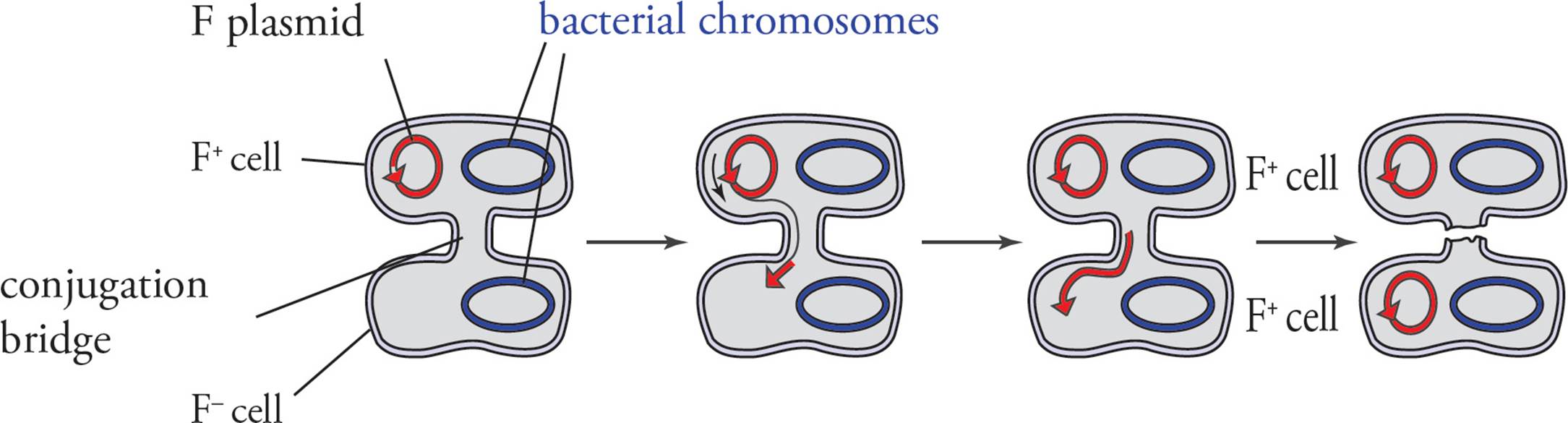
a) Conjugation and transfer of an F plasmid from an F+ donor to an F− recipient
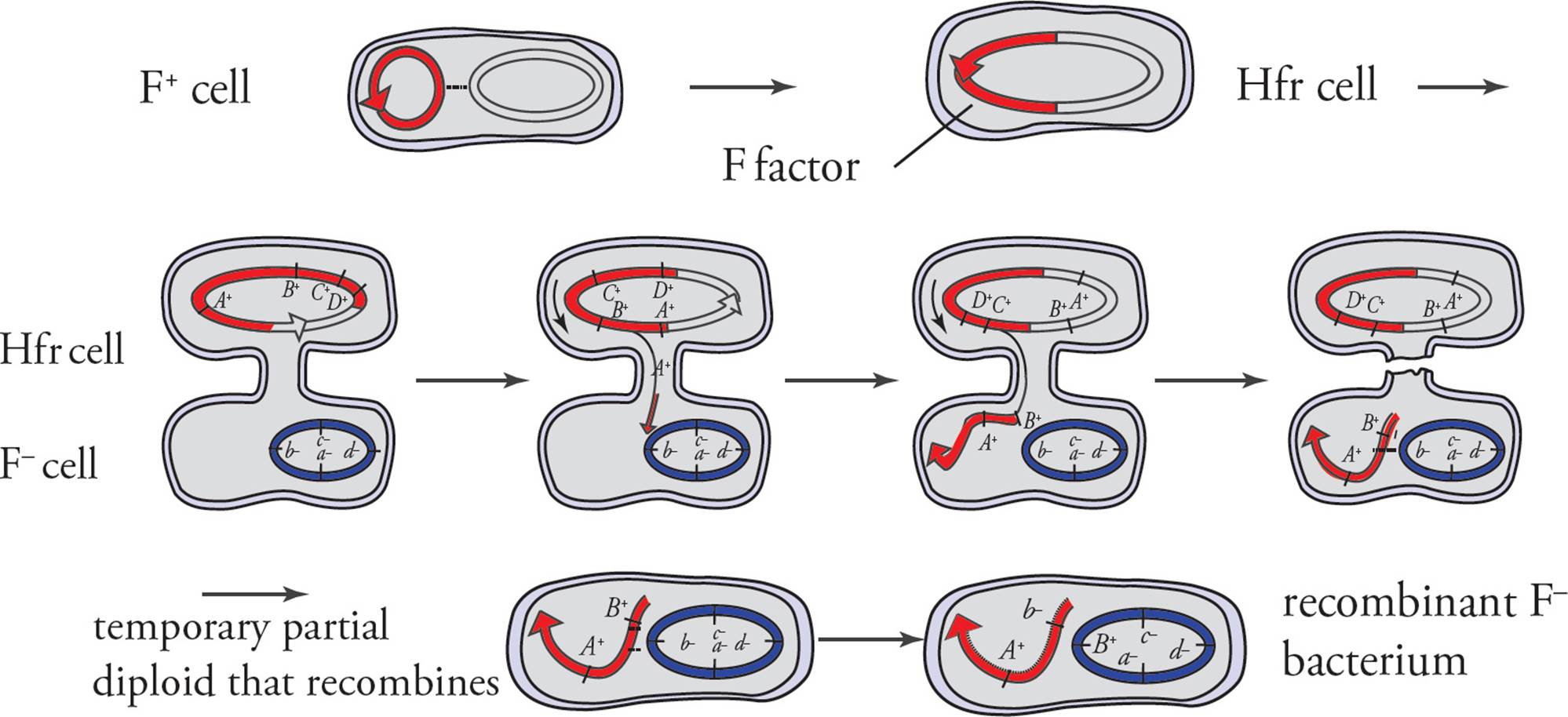
b) Conjugation and transfer of part of the bacterial chromosome from an Hfr donor to an F− recipient, resulting in recombination
Figure 13 Conjugation
Conjugation Mapping
Hfr bacteria provide a mechanism of mapping the bacterial genome. By allowing Hfr cells to conjugate in the lab and stopping the conjugation process after different time intervals, researchers can Figure out the order of the genes on the bacterial chromosome by analyzing recipient cells to see what genes were transferred.
For example, you have two strains of E. coli. One is a normal Hfr bacterium. The other is F– and auxotrophic for arginine, leucine, and histidine (F– Arg– Leu– His–). You allow conjugation to begin and stop it after 2 minutes. You find that all the recipients are now F– Arg– Leu– His+. Then you take another bunch of bacteria and allow conjugation to proceed for 5 minutes. Now all the recipients are F– Arg+ Leu– His+. You do the experiment a third and final time, allowing 8 minutes of conjugation, and find the recipients to be F– Arg+ Leu+ His+.
• What is the arrangement on the genome of the enzymes responsible for synthesis of each amino acid, relative to the site of F plasmid integration?62
Domain Archaea
Though all bacteria are prokaryotes, not all prokaryotes are equal. Certain prokaryotes belong to the domain Archaea, to be distinguished from the more “typical” bacteria (or eubacteria) which we have just discussed. The Archaea are the organisms that live in the world’s most extreme environments, including hot springs, thermal vents, and hypersaline environments (although they can also be found in less extreme environments, such as soil, water, the human colon, etc.). Structurally, they differ from other bacteria because their cell wall lacks peptidoglycan. Genetically, they share traits with eukaryotes including the presence of introns and the use of many similar mRNA sequences. However, since they are single celled, they do reproduce via fission or budding. [What does this mean for their ability to increase their genetic diversity?63]
Since Archaea have to produce enzymes that can function in extreme environments, they are of great use in industrial applications, such as food processing and sewage treatment. The development of applications for products from these cells is an ongoing area of research.
Parasitic Bacteria
Parasitic bacteria can either be obligate, meaning that they must be inside a host cell to replicate, or facultative, meaning that they can live and replicate inside or outside of a host cell. In either case, the designation as a parasitemeans that damage is being done to the host cell. However, in order to ensure a continued supply of energy and cellular materials needed to survive and reproduce, parasitic bacteria need to modulate the course of that damage. [How is this model similar to viruses?64]
T cells (lymphocytes involved in immunity, see Chapter 10) are responsible for monitoring cellular contents; people who are T cell deficient have a hard time fighting off these types of bacterial infections, just as they would also struggle with viral infections. Mycobacteria, the genus of bacteria which encompasses the cause of tuberculosis as well as other diseases, has members which are obligate and others which are facultative, whereas the sexually transmitted disease chlamydia is caused solely by an obligate parasitic bacteria.
Symbiotic Bacteria
Symbiotic bacteria coexist with a host, where both the bacterial cell and the host cell derive a benefit. An example of this would be the Rhizobia genus, which is responsible for the fixing of nitrogen in the nodules that exist on the roots of legumes. Without these bacteria the legume plants would not be able to grow, as they would be unable to derive the necessary nitrogen from the soil on their own. Similarly, Cyanobacteria are responsible for nitrogen fixing in marine environments. Some of the bacterial flora in the human gut is also composed of symbionts which aid the human body in defending against other pathogenic strains. [What other functions do the bacteria in the gut have?65] Due to their close relationship with their host cells, these bacteria often have smaller genomes with a more limited number of cellular products that are made, since the host cells can provide some of what the bacteria need. This can often mean that the symbiotic bacteria do not survive long outside of the host environment.
6.4 FUNGI
Fungal Structure
Most fungi are nonmotile, multicellular eukaryotes. One exception is yeast, which are unicellular. (Fungi possess all of the features that distinguish eukaryotes from prokaryotes, like nuclei, but for the moment we will focus on those features unique to fungi. The generalized structure of a eukaryotic cell will be discussed in Chapter 7.) The molds and fleshy fungi such as mushrooms are multicellular. All fungi possess a rigid cell wall composed of chitin, which is different from plant and bacterial cell walls but is found in the exoskeleton of insects. All fungi are chemoheterotrophs. Most are either saprophytes (feed off dead plants and animals) or parasites (feed off living organisms, doing harm to the host). Some are mutualists (live in a symbiotic relationship where both organisms benefit). For example, lichen are not plants, but rather consist of a fungus and an algae living in mutualistic symbiosis.
Most fungi are obligate aerobes, although yeasts are facultative anaerobes. The method of nutrition used by fungi is termed absorptive. This means that digestion of nutrients takes place outside the fungal cell. Simple organic molecules are then absorbed across the fungal cell wall. [Can yeast use carbon dioxide as a carbon source?66]
The fundamental fungal structure is, of course, the cell. The next level of structure in multicellular fungi is the hypha (plural: hyphae), which is a long filament of cells joined end-to-end. In septate hyphae, the cells are separated by walls called septae. Aseptate hyphae are composed of cells joined together in a long tube, in which the cytoplasmic contents and the nuclei are shared among the many cells making up the hypha. In other words, these fungi are multinucleate. Hyphae that are specialized to digest and absorb nutrients in a parasitic fashion are called haustoria (see Figure 14).
A meshwork of hyphae is called a mycelium. A large fungal structure which is visible to the naked eye is a thallus, meaning body (plural: thalli). The vegetative portion of the thallus is involved in obtaining nutrients, and the fruiting body functions in reproduction. The role of the fruiting body is to make spores. Mushrooms are fruiting bodies. The spores of fungi are distinct from bacterial endospores, both in structure and function. Each fungus may produce a great number of spores, while a bacterial cell can produce only a single endospore. [Which plays a role in reproduction, a bacterial endospore or a fungal spore?67]
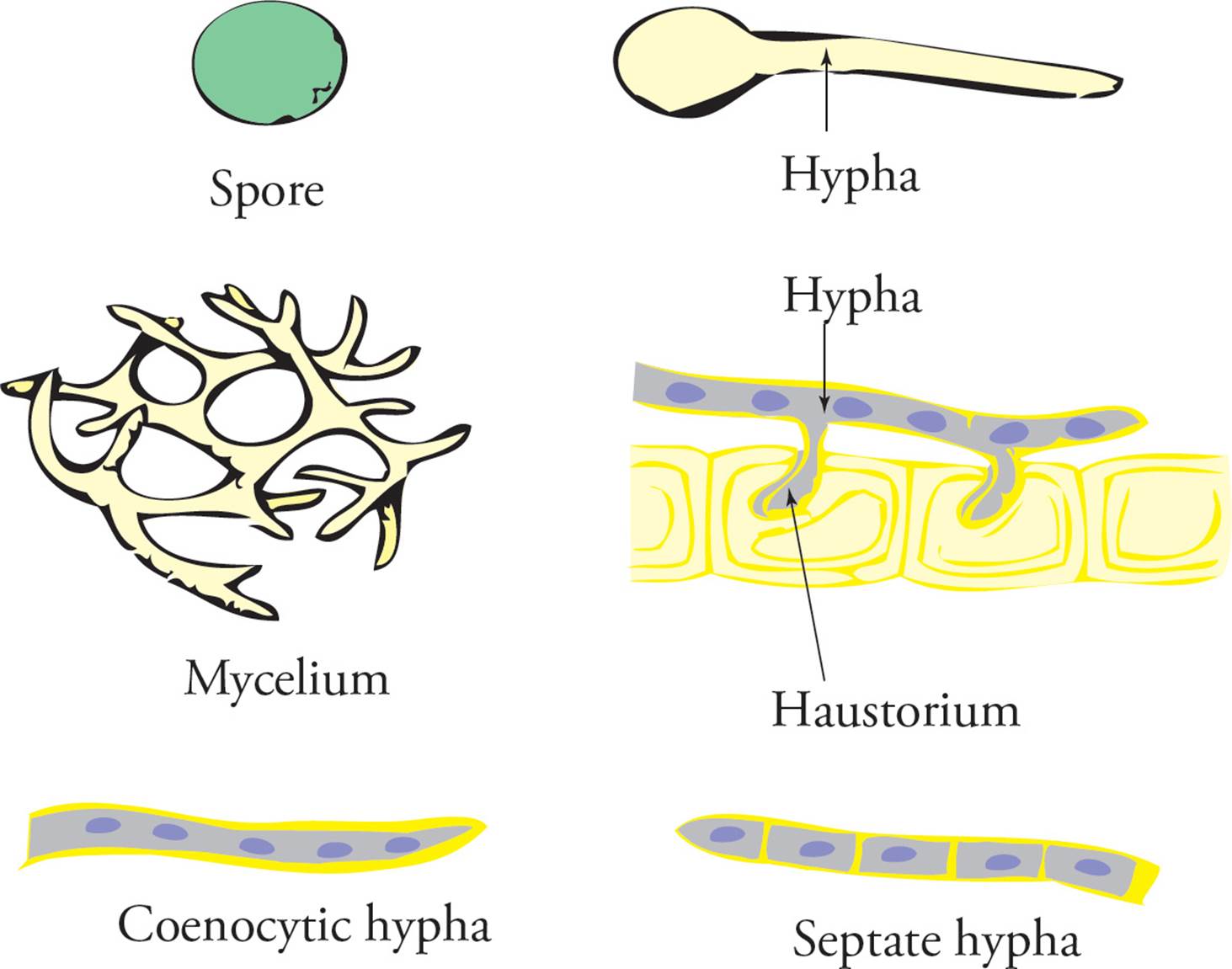
Figure 14 Some Fungal Structures
Fungal Life Cycles
Asexual Reproduction
Fungi reproduce both sexually and asexually. Asexual reproduction of fungi can occur either by budding, fragmentation, or spore production. In budding, a new smaller hypha (or single cell) grows outward from an existing one. In fragmentation, the mycelium can be broken into small pieces, each of which develops itself into a separate mycelium. Asexual spore formation in fungi occurs through mitosis to generate many spores from one cell. Spores are often produced in specialized structures, such as sporangia, found elevated on a stalk-like hypha (see Figure 15). The spores of fungi are in some cases surrounded by a tough wall resistant to environmental extremes. When environmental conditions are favorable, spores will germinate to form new hyphae. Whether budding, fragmentation, or spore production is used to reproduce asexually, the result is multiple identical genetic clones of the original fungi.
Fungal Sexual Reproduction
Every organism carries a distinct number of different (nonhomologous) chromosomes in each of its cells. Humans, for example, have 23 different chromosomes. A cell or species with only one copy of each chromosome is haploid, while those having two copies are diploid. Humans are diploid, with every somatic cell having two copies of each chromosome (leaving out sex chromosomes for the moment). There is a haploid stage in the human life cycle, however: the gamete (ova or sperm). Haploid gametes are produced from diploid cells through meiosis. Fusion of two human gametes results in a diploid zygote which then divides through mitosis to produce a diploid adult.
The life cycle of fungi is quite different. Fungal adults are haploid rather than diploid. Fungal sexual reproduction involves the fusion of haploid cells derived from haploid adults. Fusion of the haploid fungal gametes produces a diploid zygote, as in humans. In fungi, however, the diploid zygote quickly enters meiosis to produce haploid cells once again. These haploid cells produced by meiosis in fungi are not gametes but will repeatedly divide by mitosis to produce a new haploid adult.
Just as parts of fungi may specialize to reproduce asexually, some regions in fungal hyphae specialize to reproduce sexually (Figure 15). These specialized regions are termed gametangia. Gametangia can either produce and release gametes to fuse with other gametes, serve as a site for gamete fusion, or fuse with gametangia from other fungi of the same species. In some cases, fusion of the gamete nuclei does not occur right after the two haploid gametes join together. The result is a cell with two nuclei, called a dikaryon. As soon as the nuclei of the dikaryon do fuse, however, the cell enters meiosis and produces haploid cells.
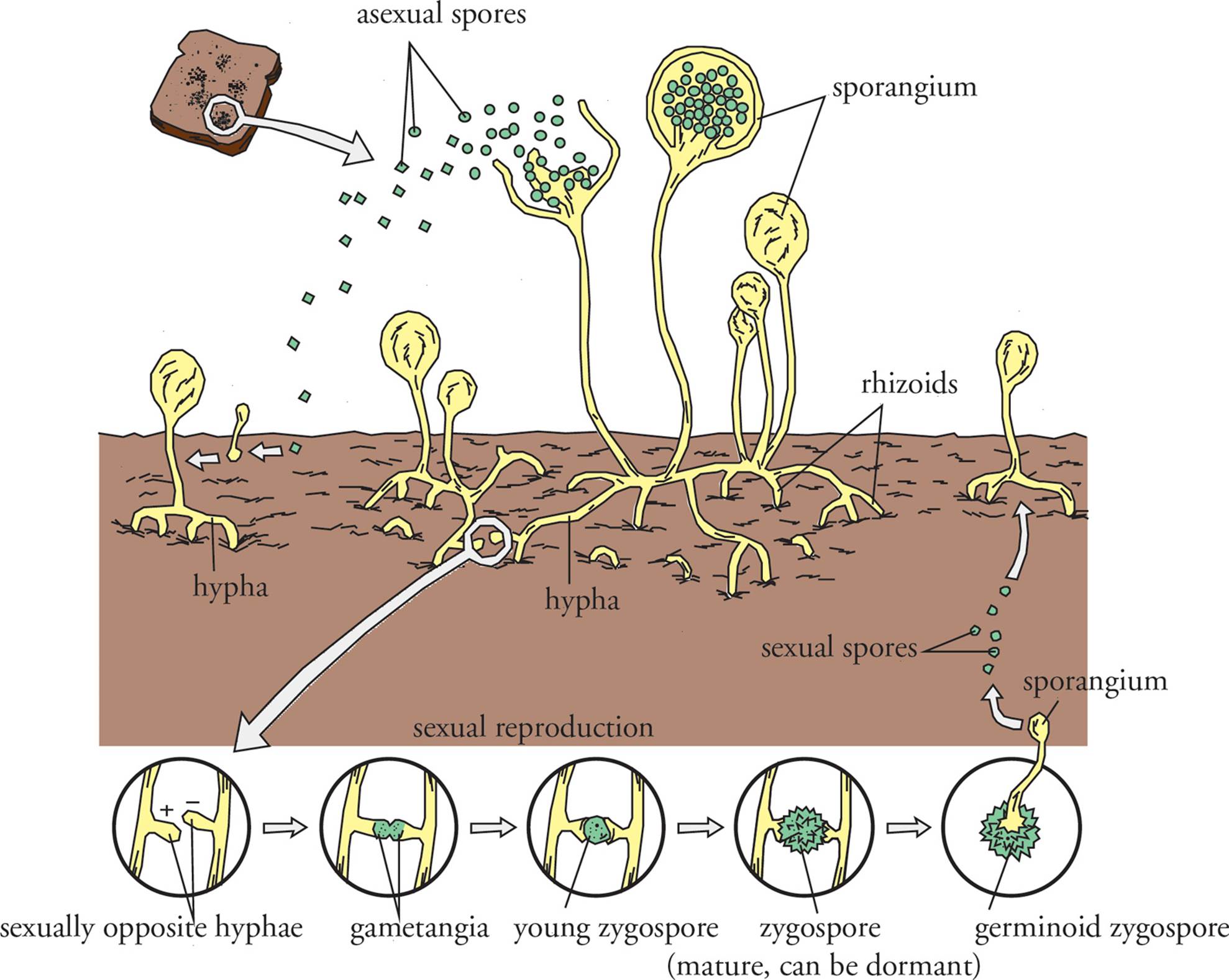
Figure 15 Fungal Reproduction
• For each item listed below, classify a disease agent which possesses the item as bacteria, virus, or fungi. (Note: More than one classification will apply in some cases.)68
1) A nuclear envelope
2) An RNA genome
3) Genes with introns
4) Ribsomes
5) Meiotic cell division
6) RNA-dependent DNA polymerase
7) Krebs cycle
8) DNA-dependent DNA polymerase
Chapter 6 Summary
• All viruses are made up of nucleic acids (either RNA or DNA) surrounded by a protein coat (capsid). They are obligate intracellular parasites and must rely on other cells to reproduce.
• Animal viruses may also have an envelope (lipid bilayer) surrounding the capsid. The envelope is derived from the host cell and is acquired by budding through the host cell membrane.
• Viral infection is specific; molecules on the viral surface determine which type of host cell it will infect.
• Viruses replicate via two major life cycles, the lytic cycle (in which more virus is made very quickly) and the lysogenic cycle (in which the virus goes dormant by integrating into the host cell genome). Viruses in the lysogenic cycle can excise from the genome and enter the lytic cycle.
• Animal viruses can also participate in a third life cycle, the productive cycle. This is very similar to the lytic cycle, but the new viruses escape by budding instead of by lysing the host.
• Lysogenic viruses can take pieces of the host DNA with them when they excise and transfer it to the next host. This is called transduction.
• RNA viruses require special virus-derived enzymes (RNA dependent RNA polymerases) in order to replicate their genomes.
• Prions and viroids are subviral particles that can cause disease and infection. They are unique in that prions are simply abnormal proteins (with no genetic material) and viroids are small pieces of RNA with no associated capsid, that do not code for proteins.
• The primary difference between prokaryotes and eukaryotes is that prokaryotes have no membrane-bound organelles (e.g., nucleus, mitochondria, etc.), thus all cellular processes occur in the cytosol.
• The shapes of bacteria can be used to classify them (round = coccus, rod = bacillus, spiral = spirochete).
• Bacteria have cell walls made out of peptidoglycan that can bind crystal violet (a purple stain used in Gram stain). Gram positive bacteria have thick cell walls and stain a dark purple. Gram negative bacteria have thinner cell walls and an outer membrane; they stain a light pink.
• Some bacteria can be classified by the presence or absence of flagella. Bacterial flagella are used for motility and are distinct from eukaryotic flagella in structure.
• Preferred growth temperature, nutrition, and oxygen use/tolerance are means of characterizing bacteria and can be used to select for growth of a particular bacteria.
• Binary fission is a means of asexual bacterial reproduction that increases the population size exponentially, but does not increase the genetic diversity of the population.
• Conjugation is a means of increasing genetic diversity in a bacterial population by exchanging DNA (plasmid or genomic) via a conjugation bridge.
• Bacteria in Domain Archaea are sometimes classified as extremophiles because they can live in harsh, extreme environments, like hot springs, thermal vents, extremes acids/bases, and hypersaline environments.
• Parasitic bacteria can live inside or outside of host cells and harm the host cells. Symbiotic bacteria coexist with host cells, but provide the host cells with a benefit, for example, the gut bacteria provide us with Vitamin K.
• Fungi are eukaryotic, have a cell wall made of chitin, and can reproduce both asexually and sexually. Yeast are unicellular fungi.
CHAPTER 6 FREESTANDING PRACTICE QUESTIONS
1. A researcher has an agar plate covered with a lawn of E. coli. She adds a drop of a substance, and the next day there is a clear spot on the plate where the substance was added. This substance could be:
I. a virus undergoing the lytic cycle.
II. a virus undergoing the productive cycle.
III. a chemical that is toxic to prokaryotes.
A) I only
B) III only
C) I and III
D) I, II and III
2. A lab technician grows a liquid bacterial culture overnight, in media without any antibiotics. The next morning, the culture is cloudy. She takes a small amount of this culture and puts it into new media containing tetracycline. The next day, she checks the culture and the media is not cloudy. What happened?
A) The bacterial culture grew the first night but not the second night.
B) The bacteria were resistant to the antibiotic tetracycline.
C) The bacteria were in the lag phase after the first night of growth.
D) The bacteria were in the stationary phase after the second night of growth.
3. Which of the following is associated with prokaryotes and does NOT introduce new genetic material?
A) Mitosis
B) Binary fission
C) Transformation
D) Transduction
4. Which of the following statements concerning viruses is true?
A) The productive cycle is the most efficient infective cycle for phages.
B) Viruses that infect human cells must have an envelope.
C) Genetic information can be transferred between hosts via transfection.
D) A virus with an RNA genome must code for an RNA-dependent RNA polymerase.
5. A researcher is trying to characterize a novel prokaryotic organism that has been found in the Indian Ocean. When Gram stained, the cells are a light pink color under the microscope. When exposed to antibiotics commonly used in the lab, the bacteria are able to enter the log growth phase in a manner similar to E. coli grown in media lacking ampicillin. A reasonable explanation is that:
A) this is a Gram-positive bacterium with an additional lipopolysaccharide layer that increases their resistance to antibiotics.
B) this is a Gram-positive bacterium with a cell membrane outside its peptidoglycan layer that increases their resistance to antibiotics.
C) this is a Gram-negative bacterium with an additional lipopolysaccharide layer that increases their resistance to antibiotics.
D) this is a Gram-negative bacterium with a peptidoglycan layer outside the cell membrane that increases their resistance to antibiotics.
6. Which of the following is true regarding prokaryotic flagella?
A) It is the predominant form of bacterial locomotion.
B) It is made of microtubules connected by dynein proteins.
C) It allows viruses to maneuver between host cells.
D) It can only be located on one end of a bacterium and this defines the polarity of the cell.
7. In prion diseases like Creutzfeldt-Jakob disease (CJD), the characteristic misfolded proteins are notoriously resistant to degradation. As a result, abnormal proteins accumulate in the endosomes and lysosomes of the cell, eventually leading to cellular dysfunction and death through a chronic, neurodegenerative process. Which of the following is the most likely explanation for the unusual resistance of these proteins to breakdown?
A) Accelerated rate of protein biosynthesis leads to early cell lysis
B) Excessive amount of protein accumulation results in cellular dysfunction
C) Aberrant protein function leads to disruption of normal cellular processes
D) Abnormal protein secondary structure results in poor binding with innate lysosomal proteases
CHAPTER 6 PRACTICE PASSAGE
Laboratory tests are a useful diagnostic tool for determining the cause of illness. One such test is the complete blood count or, CBC, as it is commonly called. In an infected individual, the white blood cell count can increase from a normal range of 4000–10,000 cells/μL to 15,000 to 20,000 cells/μL. Circulating neutrophils have a short lifespan upon release from the bone marrow (generally about ten hours); however, the demand for phagocytic cells during an infection increases markedly. The result is the release of immature neutrophils called band cells. In a differential white blood cell count, the presence of band cells is referred to as a shift to the left. A decrease in the number of neutrophils (neutropenia) can also occur as a result of inflammation or severe infection, when the removal of the neutrophils from the circulation outpaces their production. Neutropenia is also seen in certain blood cancers, such as leukemia and lymphomas, as the neutrophil precursor cells are crowded out by the cancerous cells.
The type of microorganism responsible for an infection can often be determined by changes identified in the population of white blood cells. For example, an increase in neutrophils is commonly seen in bacterial infections. An increase in eosinophils frequently accompanies parasitic infections, as well as allergic responses. A decrease in neutrophils with an increase in lymphocytes (lymphocytosis) can signify a viral infection. All types of infections can result in inflammation (fever, swelling, redness, pain).
One busy spring Saturday evening in a hospital emergency room, several patients presented with respiratory complaints. The patients had either a productive (mucus-producing) or nonproductive cough. All patients presented with some form of fever, either mild or severe. A CBC was ordered on each patient. In addition, a blood sample was obtained from each patient for the purpose of culturing and identifying any infection-causing bacteria. Standard growth media (growth media that contains glucose, amino acids, and some vitamins) providing sufficient nutrients for a wide range of bacteria was used for this purpose. The following results were obtained:
|
Patient |
Growth in culture |
|
1 |
No |
|
2 |
Yes |
|
3 |
No |
|
4 |
Yes |
Table 1 Bacterial Culture Results in Four Different Patients
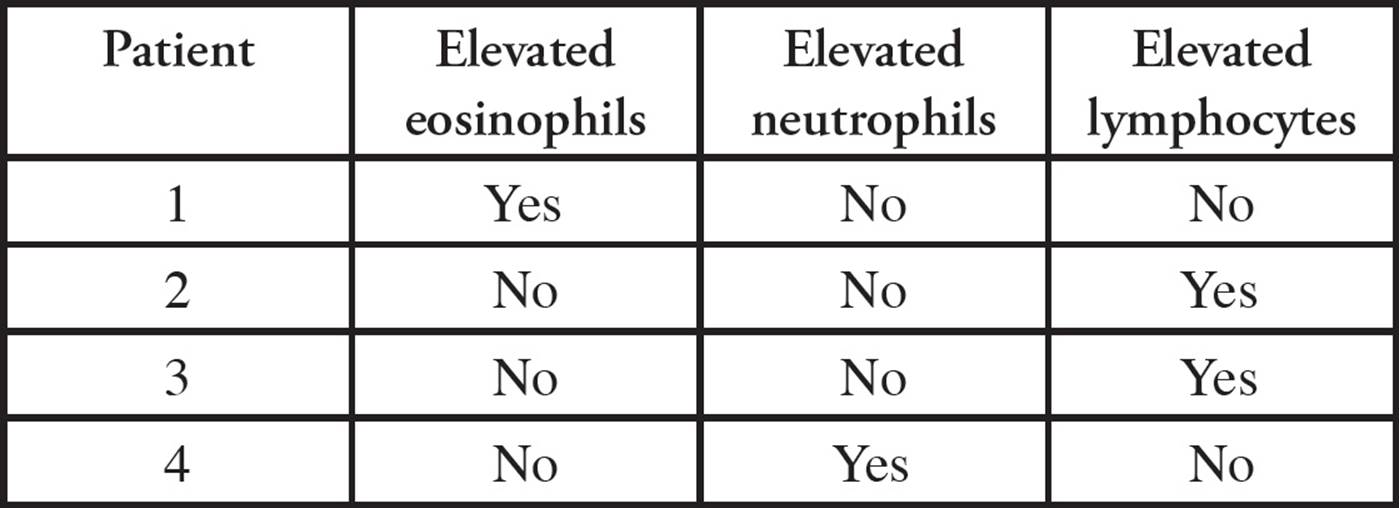
Table 2 WBC Counts in Four Different Patients
1. If placed on a course of antibiotic therapy, which of the following patients would feel significantly improved after approximately 1–2 days?
A) Patient 1
B) Patient 2
C) Patient 3
D) Patient 4
2. Why did the culture performed on the sample obtained from Patient 3 not yield any growth?
A) The bacteria causing the infection in Patient 3 is a uracil auxotroph.
B) Patient 3 has a bacterial infection.
C) Patient 3 is suffering from allergic symptoms.
D) A different growth medium was required.
3. Patient 4 was prescribed a broad-spectrum antibiotic and released. The patient returned to the emergency room after a two-week period complaining of worsening symptoms. The patient admitted to discontinuing use of the antibiotic after four days of therapy because they felt much improved. Which of the following are possible explanations for the patient’s symptoms?
I. The susceptible bacterial population was not fully eradicated.
II. The patient was resistant to the antibiotic.
III. A resistant population of bacteria has begun to proliferate.
A) I only
B) II only
C) I and III only
D) I, II, and III
4. To determine the most appropriate type of antibiotic to prescribe, which of the following additional tests could be performed on a patient sample for classification purposes?
A) Phage-typing
B) Gram-staining
C) Fermentation
D) Transduction
5. Which of the following bacterial types are LEAST likely to cause a respiratory infection?
A) Tolerant anaerobe
B) Obligate aerobe
C) Facultative anaerobe
D) Obligate anaerobe
6. If a patient’s symptoms included neutropenia and elevated lymphocyte counts, which of the following diagnoses could be possible?
I. Allergies
II. Leukemia/lymphoma
III. Viral infection
A) I only
B) II only
C) I and III only
D) II and III only
7. An experimental therapy to treat patients with multiple antibiotic-resistant bacteria involves introduction of a highly specific bacteriophage to the infected patient’s bloodstream. Which of the following bacteriophage types would be the LEAST useful for this type of therapy?
A) A lytic bacteriophage
B) A lysogenic bacteriophage
C) An RNA virus
D) An enveloped virus
SOLUTIONS TO CHAPTER 6 FREESTANDING PRACTICE QUESTIONS
1. C A clear spot on a plate (known as a plaque) indicates that the E. coli are dead. This could be due to the addition of a lytic virus (Item I is true and choice B can be eliminated) or toxin (Item III is true and choice A can be eliminated). However, only animal viruses can go through the productive cycle because viruses cannot bud out of a cell with a cell wall, such as bacteria (Item II is false; choice D can be eliminated and choice C is correct).
2. A Cloudy cultures are usually in the stationary phase and clear cultures are either not growing or still in the lag phase. Since the culture was cloudy on the first morning, bacteria had grown overnight and were most likely in stationary phase (choice A is correct and choice C is wrong). The culture on the second morning was clear, indicating minimal growth (choice D is wrong). Since the first overnight culture did not contain tetracycline and the second overnight culture did, it is possible that the strain was sensitive to tetracycline, not resistant (choice B is wrong).
3. B Binary fission is the means by which bacteria divide and reproduce. It produces two progeny cells that are genetically identical to the parent; no new genetic information is introduced (choice B is correct). Although mitosis also does not introduce new genetic information, it is a process undergone by eukaryotic cells, not prokaryotes (choice A is wrong). Both transformation and transduction are associated with prokaryotes, but both involve the introduction of new genetic material. Transformation is the uptake of genetic material (plasmids or chromosomal DNA) from the extracellular environment (choice C is wrong) and transduction is the transfer of genetic information from one bacteria to another via a lysogenic phage (choice D is wrong).
4. D In order to replicate its genome, an RNA virus must code for an RNA-dependent RNA polymerase; this enzyme will create a new strand of RNA by reading a template strand of RNA. Viral host cells will not express these enzymes naturally; they have no need to make RNA by reading RNA. Host cells normally produce RNA using DNA as a template (choice D is correct). Phages only infect bacteria, and can only undergo the lytic and lysogenic cycles; the productive cycle involves budding through cell membrane and cannot occur in hosts with cell walls, such as bacteria (choice A is wrong). Although viruses with an envelope (lipid bilayer coating) are restricted to infecting animal cells, the outer membrane is not required (choice B is wrong). Genetic information can indeed be transferred between hosts, but this process is called transduction, not transfection (choice C is wrong).
5. C The answer options all start with Gram positive or Gram negative, and the light pink staining in the question stem indicates that this bacteria is Gram negative. Gram-positive bacteria have a peptidoglycan layer outside the cell membrane and therefore stain a dark purple (choices A and B are wrong). Gram-negative bacteria have a lipopolysaccharide layer outside the peptidoglycan layer. This additional outer layer prevents dark staining (hence the light pink color) and increases resistance to antibiotics (choice C is correct and choice D is wrong).
6. A Prokaryotic flagella are the predominant means of bacterial locomotion (choice A is correct). Only eukaryotic flagella are made of microtubules and dynein; bacterial flagella have a different structure and are made of the protein flagellin (choice B is wrong). Viruses rely on diffusion to maneuver between host cells, not flagella (choice C is wrong). Bacteria can have flagella on one end (monotrichous), both ends (amphitrichous), or in multiple places (peritrichous; choice D is wrong).
7. D Typically in a cell, abnormal proteins are targeted to proteasomes in the cytosol for degradation or to the lysosomes for digestion. The accumulation of the abnormal prions in the lysosomes suggests that they are somehow resistant to the lysosomal enzymes, and this could be due to their abnormal structure that prevents efficient binding at the active sites of the lysosomal digestive enzymes (choice D is correct). CJD and other prion diseases do not involve accelerated protein biosynthesis; normal prion proteins are produced at their typical rate, but are converted after translation into the abnormal prion version (choice A is wrong). While the accumulation of misfolded proteins likely contributes to cellular dysfunction, and while prions could (and likely do) lead to disruption of normal cellular processes, neither of these explain why the abnormal proteins are resistant to breakdown (choice D is a better choice than either B or C).
SOLUTIONS TO CHAPTER 6 PRACTICE PASSAGE
1. D Antibiotics only treat bacterial infections. Table 1 shows that Patients 2 and 4 have positive cultures, confirming a bacterial infection (choices A and C can be eliminated). However, from Table 2, only Patient 4 has elevated neutrophils, which the passage states is indicative of a bacterial infection, making this patient a candidate for antibiotic therapy. Patient 2 has elevated lymphocytes (not neutrophils), which indicates a concomitant viral infection; the lack of elevated neutrophils in this patient is most likely the result of overwhelming infection. While the bacterial infection of Patient 2 would begin to subside by 1–2 days, the viral infection would take longer to eradicate, and therefore this patient would still be feeling poorly at this point (choice D is better than choice B).
2. D From Table 2, Patient 3 has elevated lymphocytes, indicating a viral infection (choices A, B, and C can be eliminated); no growth would occur on standard growth media. Viruses are obligate intracellular parasites and require special growth media containing live cells for reproduction. According to the passage, standard growth media for bacterial culturing was used. Note that if the patient had elevated neutrophils and normal lymphocyte levels (indicating a bacterial infection), choice A would be the best answer, since uracil was not added to the culture media.
3. C Item I is true: If the symptoms return and/or worsen, it is possible that the infection was not completely eradicated (choice B can be eliminated). Item II is false: Bacteria are resistant, the patient is not (this is a common misconception; choices B and D can be eliminated). Item III is true: It is also possible that a strain resistant to the antibiotic being used remained alive and is now proliferating (choice C is correct).
4. B Determination of Gram status can aid in antibiotic selection. Phage typing applies to bacteriophage, not bacteria (choice A can be eliminated). Fermentation refers to metabolic activity (choice C can be eliminated). Transduction is not a classification method (choice D can be eliminated).
5. D The lungs are a high oxygen environment that is unfavorable to obligate anaerobes. Tolerant anaerobes, obligate aerobes, and facultative anaerobes can all survive in the presence of oxygen (choices A, B, and C can be eliminated).
6. D Item I is false: The passage states that elevated eosinophil counts (not lymphocytes) accompany allergic reactions (choices A and C can be eliminated). Note that both remaining answer choices include Item II, so it must be true: The passage states that neutropenia and elevated lymphocyte counts are seen in lymphoma and leukemia (blood cancers). Item III is true: neutropenia and lymphocytosis are seen in viral infections (choice B can be eliminated and choice D is correct).
7. D Enveloped viruses infect only animal cells and would not be useful in eliminating bacteria from a human patient. The most useful type of virus would attack the infecting bacteria and cause the bacteria to lyse, eradicating the patient’s infection (choice A would be useful and can be eliminated), and this virus could have either an RNA or a DNA genome (choice C could be useful and can be eliminated). A virus that incorporates itself into the bacterial genome and then goes dormant (a lysogenic virus) would not be as helpful as a lytic virus in eradicating an infection; however, it would still be more useful than a virus that cannot infect bacteria at all (choice B would be more useful than choice D).
1 Note, however, that some viruses store some ATP in their capsids. They acquired this ATP from the previous host and typically use it to power penetration (see below).
2 The mammalian cells are directly dependent on the ATP generated by the electron transport chain, so if cyanide inhibits the electron transport chain, the mammalian cells will die (choice D is wrong). The viruses are dependent on the mammalian cells for the ATP and enzymes needed for replication, so if the mammalian cells die, viral replication will stop (choice C is wrong). However, any viruses that had already completed the replication process when the cyanide was added will not be affected, and will remain infectious (choice B is correct and choice A is wrong).
3 There are exceptions. For example, it has recently been discovered that the Hepatitis B virus has a circular DNA genome which is part single-stranded and part double-stranded. The take-home point here is that when a virus is not inside a host cell, it contains only its genome, which is always the same (except in special situations such as when a piece of host genome accidentally becomes incorporated in the viral genome). In contrast, a true cell contains not only its genome, but also mRNA, rRNA, and tRNA.
4 Adenine base pairs with thymine in double-stranded DNA. Thus, for every A there should be one T for a one to one ratio of A to T. If the ratio differs from this, the genome must be single-stranded DNA, or RNA, which has no T.
5 No, it cannot be a virus. Viruses possess only one kind of nucleic acid. The disease agent is another kind of obligate intracellular parasite (certain bacteria can only reproduce inside host cells, e.g., Chlamydia).
6 The viral genome will probably no longer fit within the normal viral structure, and the genome will therefore not be packaged into infectious viral particles.
7 Viruses use host ribosomes. Viral and host proteins are translated by the same ribosomes.
8 The problem is that the virus must contain at least 750 bp (250 amino acids) and 900 bp (300 amino acids) of genetic information for unrelated polypeptides in 1000 bp of DNA. The only way to do this is overlapping multiple reading frames (choice C).
9 Phage must puncture the bacteraial cell wall, while animal viruses can be internalized whole into animal cells (since they do not have a cell wall).
10 Remember: Viruses acquire envelopes by budding through host membranes. Phages and plant viruses infect hosts that possess cell walls. When viruses begin to exit the cell, the cell wall is destroyed, and host membranes rupture. Hence there is no membrane through which the remaining viruses must bud; they simply escape in a lytic explosion.
11 It suggests that the virus is enveloped, so the antibody cannot reach its epitope on the capsid surface in infectious virus.
12 No. The host cell would lyse before the phage had time to replicate and assemble.
13 The initial decrease is due to the simple fact that many phage have injected their genomes into hosts and are no longer infectious.
14 The 35S cysteine will be incorporated into viral coat proteins and the 32P phosphate will be incorporated into the viral nucleic acid genome in newly released viral particles. (Proteins contain no P and nucleic acids contain no S.) When these viruses infect bacteria, their nucleic acids are injected into the bacteria while the capsid proteins remain on the exterior, which means that only the 32P will be found in the interior of the newly infected cells.
15 When two viruses infect the same cell, it is called co-infection. Some normal viruses will result, and some genomes from defective viruses will get packaged into capsids made from proteins encoded by the normal virus. The latter will be capable of infecting new hosts, but when they do their progeny will not survive due to the capsid abnormality. Choice A is correct and choices B and D are wrong. Think about it: where did the phage with the deleted capsid gene come from?! The deficient virus must have come from a co-infection such as this. The deficient phage can only infect host cells and reproduce with the help of normal viruses. Note that because a single virus carries only a single genome, it can produce only one type of progeny (choice C is wrong).
16 Prophage latency results from a viral repressor protein binding to viral DNA in a sequence-specific manner. The specific DNA sequence to which the repressor binds is present in the viral genes but not in the bacterial genes, so the bacterial gene can be expressed while the viral genes are repressed.
17 Yes, it would. The soluble CD4 protein would bind to the virus’s CD4 receptor (gp120) and block attachment of the virus to the T cells.
18 Two reasons: 1) The receptor has a specific role in the normal physiology of the host, which a mutation might compromise. 2) Viruses generally evolve so rapidly that they can keep up with any changes in the host, but this is not an absolute rule. Cells of our immune system keep us alive by keeping up with most microorganisms’ tricks.
19 The detergent solubilized the viral envelope (choice C is wrong). As stated in the text, some envelop proteins are encoded by the virus and some are derived from the host’s membranes during budding (choice A is correct and choice D is wrong). Removal of envelope proteins will impair viral adsorption and reduce infectivity (choice B is wrong).
20 Bacteria do not perform endocytosis, in part because they have a rigid cell wall which does not permit them to.
21 to copy the RNA genome for viral replication; the host never makes RNA from RNA.
22 No. The (+) strand RNA virus will be able to produce viral genome and proteins, but progeny will not be able to acquire the envelope they need to be infectious.
23 To make (+) strand copies of the genome, the virus needs the complementary strand as a template: the (–) strand RNA. Thus, the RNA-dependent RNA polymerase produces a (–) strand intermediate before generating new (+) strand genomes.
24 Neither. Viral RNA-dependent RNA polymerase first makes (+) strand as mRNA and then uses the (+) strand as the template to replicate new (–) strand genomes.
25 Because the viral RNA genome can be translated by host ribosomes; thus reverse transcriptase may be made after the viral genome enters the host. It just so happens that HIV does carry its reverse transcriptase within its capsid. You should understand why this is not a theoretical necessity.
26 No, it will not. Reverse transcriptase is required for only one phase of the retrovirus life cycle: the copying of the viral RNA genome into DNA so that it can integrate into the host genome and be transcribed. Once the viral genome has integrated, transcription to produce viral mRNA and new viral RNA genomes does not involve reverse transcriptase. It can proceed with the normal host-cell enzymes.
27 The host cell will only make dNTPs in preparation for replication. If the virus wants to reproduce without waiting for the host to do so, it must encode its own enzymes for the synthesis of DNA building blocks.
28 Transcription is always occurring in all cells, so NTPs (not dNTPs) are always present.
29 The error rate in RNA synthesis is much higher than in DNA synthesis, in part because there are mechanisms to proofread and correct errors in DNA synthesis (but not in RNA synthesis). If an RNA genome were too large, every copy of the viral genome synthesized would suffer from so many errors that no infectious virus would be produced.
30 To replicate, the DNA virus must either provide all of the necessary components (such as dNTPs) itself, infect a cell that is already dividing, or induce the cell it infects to enter mitosis and produce the ingredients for DNA synthesis.
31 Yes, thanks to complementation. The mutant viruses will complement each other, one providing the E1A protein and the other providing DNA polymerase. Note that this had to have happened before; how else could a defective virus such as these exist? One virus which complements another is called a helper virus.
32 The Central Dogma states that information flows in its nucleotide form from DNA to RNA (transcription), and then in its amino acid form from RNA to protein (translation). Prions take both transcription and translation out of the process and have proteins being shaped based on other proteins, hence the term “self-replicating”.
33 There are a few exceptions to this (e.g., bacteria with more than one chromosome and/or linear chromosomes), but you do not have to know them for the MCAT.
34 The 5 end of the mRNA polymer is free, since elongation of mRNA proceeds 5 to 3. Proteins are made N to C, so the free end of the polypeptides is the N terminus.
35 It would burst, since water would flow into the cell by osmosis.
36 Lysozyme hydrolyzes linkages in peptidoglycan to weaken the cell wall. The peptidoglycan in Gram-positive cells is more accessible, since these cells do not possess an additional outer layer; therefore, Gram-positive cells will lyse more easily when treated with lysozyme.
37 No. Viruses lack any means of energy production on their own and any means of active movement. They rely on diffusion to find host cells.
38 Monotrichous
39 The capsule, or glycocalyx is also involved in adherence. And yes, fimbriae do play a role in infection, by facilitating adhesion to cells so that the bacteria can colonize a tissue.
40 Normally you expect decreasing temperature to decrease the rate of all chemical, biochemical, and biological processes, since reactions accelerate when kinetic energy increases. However, bacteria which have evolved to live at low temperature (psychrophiles) possess enzymes which may be optimally active at low temperature, leading to better growth.
41 Since there’s no sunlight in the bloodstream, B and D are out. If it’s a parasite, it most likely uses some of our chemicals, so it must be a heterotroph, which eliminates A. The answer is C.
42 The ability to use sunlight indicates that the organism is a phototroph, and the inability to use carbon dioxide as a carbon source indicates that it is a heterotroph—it must use organic molecules as a carbon source. The answer is D.
43 Sixteen times as much (refer to Chapter 4).
44 The enzymes include superoxide dismutase (converts O2– to H2O2) and catalase (converts H2O2 to H2O + O2). An example of a harmful O2 by-product is superoxide anion, O2–.
45 The bacterium cannot be a facultative anaerobe, since the question states it cannot use O2. It could be either an obligate or a tolerant anaerobe depending on its ability to neutralize harmful oxygen free radicals.
46 Since the bacteria grew only at the bottom of the tube, farthest away from any oxygen, this indicates that they could only grow in the absence of oxygen. Thus, they are obligate anaerobes.
47 Because NAD+ must be regenerated from NADH for glycolysis to continue. In fermentation, the electrons are passed from NADH to a molecule other than O2, such as pyruvic acid.
48 Aerobic respiration produces 32 ATP per glucose in prokaryotes compared to only 2 ATP per glucose in fermentation. If the rate of growth is to remain the same, the rate of ATP production must remain the same to drive biosynthetic pathways forward. Since fermentation produces 1/16 the amount of ATP per glucose, the rate of glucose consumption must increase sixteen fold to maintain the rate of growth at the same level. The answer is A. (In reality the growth rate would probably decrease.)
49 No. Each daughter cell is identical to the parent cell (assuming no mutation took place).
50 Many eukaryotes reproduce asexually. Sexual reproduction allows for generation of new allelic combinations through meiotic recombination and random union of gametes. Without this, diversity will decrease over time.
51 In eukaryotes, asexual reproduction occurs through mitosis. Prokarotes do not go through mitosis.
52 Since four hours is equal to 240 minutes, the bacteria will divide twelve times. Therefore, one bacterium will produce 212 = 4096 bacteria after 12 divisions. Since there are 10 bacteria initially, the total after four hours will be 10 × 212 = 40,960.
53 Cells that are not growing are not actively producing components that are needed for cell division, such as dNTPs. The lag period is a time when biosynthetic pathways are very actively producing new cellular components so that cells can then begin to divide.
54 No, since they will have all the gear necessary for population growth at the ready.
55 The bacteria will use glucose during the lag phase to produce ATP and cellular machinery. During this period, glucose is abundant, and the cell is actively performing biosynthesis, so glycolysis is very active. During the stationary phase, however, the glucose will be depleted, and the rate of metabolism will have slowed dramatically, so the rate of glycolysis will decrease as well.
56 Stationary. Forming an endospore is like hibernating, not reproducing. Bacteria do it in order to sleep through the bad times.
57 No. Conjugation occurs only between F+ (male) and F– (female).
58 The male cell contains the F factor that encodes the genes for pili production and will produce pili.
59 Yes. All of the genes of the F factor are still present and expressed normally in the Hfr cell.
60 When an Hfr cell conjugates with an F– cell and transfers a portion of the bacterial chromosomes, the F– cell will have two copies of some genes, and recombination can occur between the two copies.
61 Yes. Genes for proteins of related functions are often adjacent to each other in prokaryotes (in operons) and so will transfer to an F– cell together.
62 The experiments showed that the ability to make histidine was transferred in a short time. After a slightly longer time, the ability to make both histidine and arginine was transferred. Lastly, the ability to make leucine were transferred. So the arrangement on the genome (the map) must be: His-Arg-Leu-plasmid integration site.
63 Archaea would need to use separate strategies to increase their genetic diversity, just like eubacteria. The ability to become more genetically diverse would not be built into reproduction as it is in humans, in part because meiosis is not occurring.
64 Viruses are obligate intracellular parasites. They do not have the option to replicate outside of a host cell, but must also balance the damage that is done to the host cell against what is needed for more virus to be made.
65 The gut flora is responsible for the production of vitamin K, which is necessary for blood clotting and in feeding off of undigested material from what humans have consumed, they are one of the final stages in processing our solid waste for excretion.
66 No. All fungi are chemoheterotrophs, so by definition require an organic (derived from another organism) carbon source.
67 Only the fungal spore. Remember that the bacterial endospore just allows the bacterium to wait out the bad times.
68 1: fungi. 2: virus. 3: virus, fungi. 4: bacteria, fungi. 5: fungi. 6: virus. 7: bacteria, fungi. 8: bacteria, virus, fungi.