MCAT Biology and Biochemistry: New for MCAT 2015 (2014)
Chapter 8. Genetics and Evolution
The nature of the fundamental unit of inheritance, the gene, has been agreed upon by scientists only since the mid-nineteenth century. Aristotle believed that traits were passed on in the form of “pangenes,” particles derived from all parts of the body and distilled into eggs and sperm. In the seventeenth century, different theorists believed that all genetic information was passed by either the father or the mother. Finally, early in the nineteenth century, people began to see that characteristics are passed from both parents; this led to the idea that parental characteristics were evenly mixed in offspring, in a process termed “blending.” The notion that some characteristics were inherited in an either- or fashion, while others were in fact blended, remained unconceived.
The proponents of these early theories cannot be faulted for their lack of electron microscopes and other modern tools and techniques. But one is tempted to criticize their ideas for their obvious irrelevance to reality. One didn’t need a Cray supercomputer to Figure out that both parents contributed to a child’s makeup. Why did researchers fail to arrive at this seemingly obvious hypothesis? Probably for two reasons: methods and dogma.
First, their approach to discovery was not empirical, but rather a priori. They believed knowledge could be derived by speculation alone, and that to perform experiments in the physical world was to dirty one’s hands. And second, the prevailing religious dogma strongly censored empirical exploration, since it threatened the metaphysical tenets of the church.
What is different today is the approach to discovery known as the scientific method. Modern scientists know that only through careful, sober consideration of a question, formulation of a tentative answer, and testing of that hypothesis can new knowledge be uncovered. In this chapter we will examine what is now considered to be the truth about genetics and evolution, all the way back to the origin of life.
We challenge you to attack this knowledge in the way its discoverers did. This is difficult material. Spend time thinking about the in-text questions before reading the answers. If you have trouble with a topic, stop and take out a fresh piece of paper. Write down all the facets of your current understanding, and look for internal inconsistencies and fallacies. Make up your own Punnett squares, pedigrees, and sketches of chromosomes during meiosis if the ones we present aren’t sufficient. And finally, as you review, ask yourself which of the modern “truths” will one day be looked back on as preposterous ponderings of blindfolded pseudo-scientists.
8.1 INTRODUCTION TO GENETICS
Genetics is the science that describes the inheritance of traits from one generation to another. At the origin of genetics, patterns of inheritance were observed to follow certain predicTable patterns, as described by Mendel’s laws. The reasons for these patterns of inheritance were to remain a mystery until the nature of DNA as the genetic material was known. Today we can use our knowledge of DNA and the cell to understand Mendel’s laws at the molecular level.
DNA as the Genetic Information of the Cell
Gregor Mendel described the basic principles of heredity (see Section 8.3), but not the molecular foundations. It wasn’t until years later that DNA was shown to be the building block of cellular genetics. Mitosis and meiosis were actively studied in the late 19th century, and this paved the way for the chromosome theory of inheritance, which indicated that genes are located on chromosomes. Thomas Hunt Morgan associated a specific gene and its subsequent phenotype (eye color in the fruit fly) with a specific chromosome (the X chromosome). However, chromosomes are composed of both DNA and protein. It wasn’t until an important series of experiments, which started in the 1920s, that DNA was accepted as the genetic material of the cell.
First, Frederick Griffith showed that cell extracts can transform bacteria, indicating biological macromolecules carry hereditary information. This work made use of two strains of Streptococcus pneumoniae. One strain is lethal in mice and is characterized by a smooth (S) appearance under the microscope due to the presence of a polysaccharide capsule. The other strain is less virulent and does not kill mice. It lacks the capsule and so has a rough (R) appearance under the microscope. When heat-killed S bacteria were injected into mice, the animals survived. However, when heat-killed S strain was mixed with live R strain and then injected together, the mice died. This suggested that the cell extract of dead S strain was capable of conferring virulence to the R strain. In addition, it was found that live S strain S. pneumoniae could be isolated from the dead mice (Figure 1).
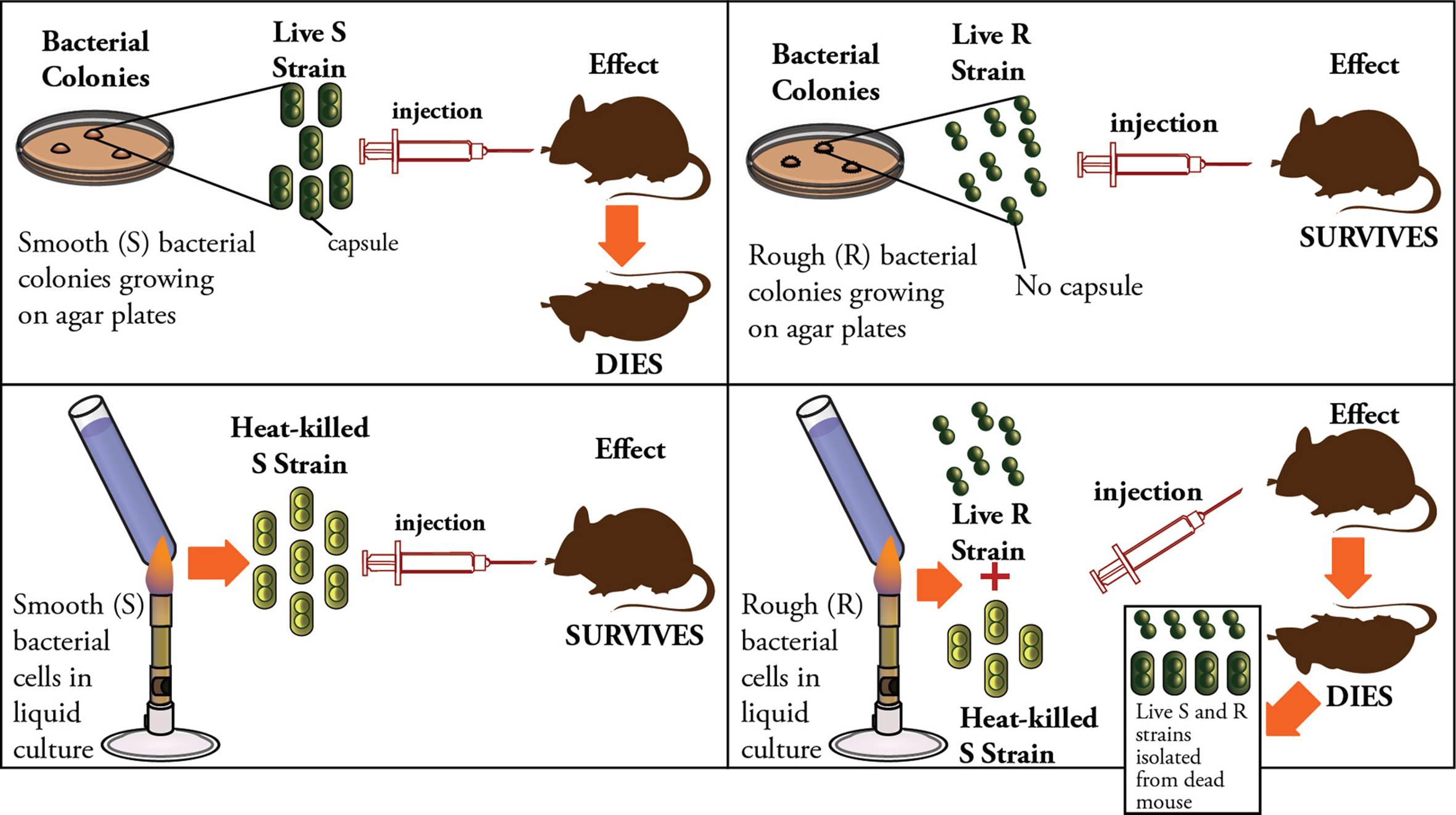
Figure 1 The Griffith Experiment
However, it still wasn’t clear how the R strain was obtaining virulence. To Figure this out, Oswald Avery, Colin MacLeod, and Maclyn McCarty systematically and chemically destroyed each biological macromolecule in the extracts from dead S strain S. pneumoniae. These treated extracts were then injected into mice with live R strain. Since the two strains were phenotypically different because of a polysaccharide coat, an obvious hypothesis was that virulence was being transferred via a polysaccharide. However, when polysaccharides were destroyed, virulence was still transferred from the dead S bacteria to the live R bacteria. Indeed, this was the case for all macromolecules save one: DNA (Figure 2). When DNase was added to S strain extracts, virulence was not conferred to the R strain. This suggested that DNA was able to transform bacteria, and was the molecule of heritability that scientists had been looking for.
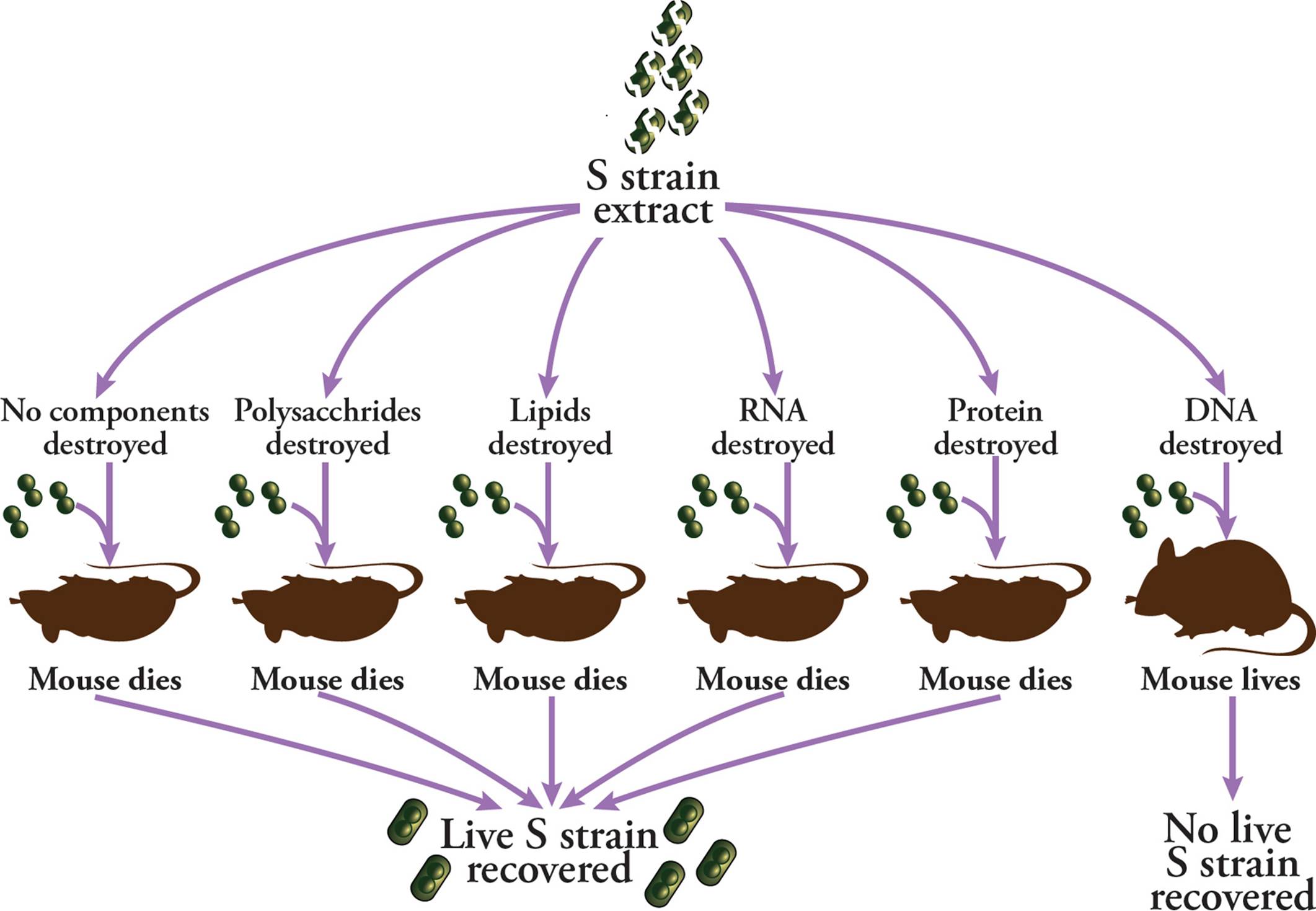
Figure 2 The Avery-MacLeod-McCarty Experiment
Still, there were scientists who didn’t believe DNA contained cellular genetic information. How could a molecule with only four monomers (A, C, G and T) be the basis of such extensive genetic diversity? Wouldn’t proteins (with twenty amino acid monomer building blocks) be more suited to this job?
Definitive experiments were done by Alfred Hershey and Martha Chase. The model organism here was the phage T2, a virus that infects bacteria. Hershey and Chase grew two parallel cultures of phages in bacterial hosts. One contained 32P, a radioactive isotope of phosphorus. Phages made in this culture contained radioactively labeled DNA, since DNA contains phosphorus in backbone phosphate groups. The second culture contained 35S, a radioactive isotope of sulphur. In this culture, phages with radioactive protein capsids were made (since amino acids such as methionine and cysteine contain sulphur atoms). The two labeled phage samples were then used to infect new cultures of unlabelled bacteria. Later, the cultures were centrifuged (spun at high speeds); this caused the bacterial cells to settle into a pellet at the bottom of the tube. The liquid layer (called the supernatant) contained growth media and phage ghosts (capsid particles without internal nucleic acids). Hershey and Chase found that for phages grown in 32P, the radioactive label was transferred to bacteria host cells in the pellet. This confirmed that DNA was being transferred and was therefore the hereditary material. 35S was not transferred to the bacterial cells, indicating that capsid proteins remained outside the host cell (in the supernatant) and therefore do not contribute to heritability.
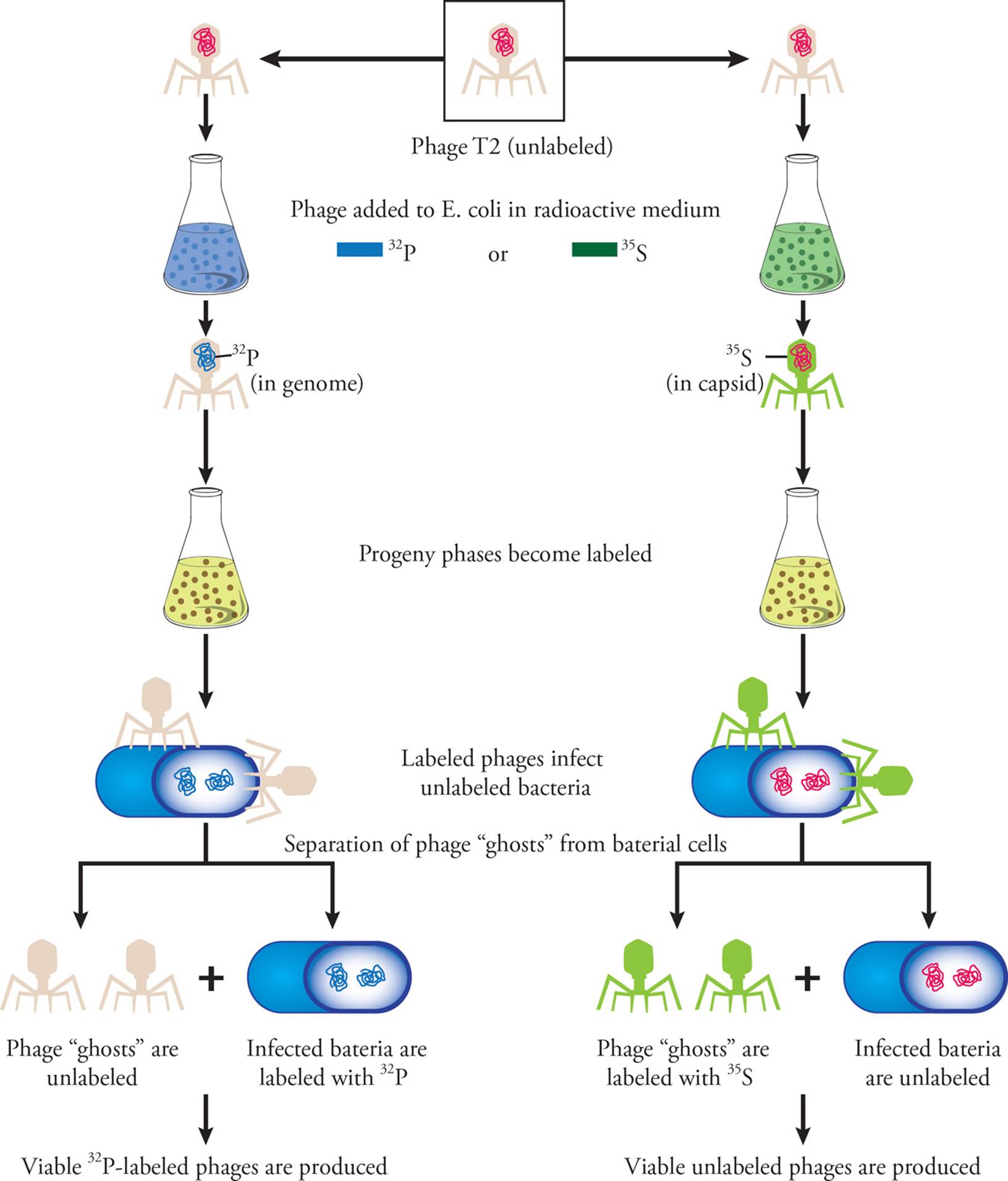
Figure 3 The Hershey-Chase Experiment
In addition to these three seminal projects, some additional evidence supports the fact that DNA carries genetic information of the cell:
• The total amount of DNA in a given cell (and species) is constant, and work by Erwin Chargaff suggested that each species has a consistent make-up of DNA; human DNA for example is 30.9% adenine, 29.4% thymine, 19.9% guanine and 19.8% cytosine.
• Matthew Meselson and Franklin Stahl showed that DNA replication is semiconservative; cellular DNA is copied during each cell cycle, and is therefore self-perpetuating and consistent (see Chapter 5).
• In quiescent (resting) cells, macromolecules such as carbohydrates and proteins have a relatively short half-life and are constantly recycled and replaced. DNA is not broken down.
• Studies involving mutagens showed that DNA-altering chemicals induce phenotypic mutations, as do wavelengths of light that are absorbed by DNA.
Genes and Alleles
One of the basic tenets of genetics is that children inherit traits from both parents. Humans have a life cycle in which life begins with a diploid cell, the zygote. Diploid organisms (or cells) have two copies of the genome in each cell, while haploid cells have one copy of the genome. In sexual reproduction, the diploid zygote is produced by fusion of two haploid gametes: a haploid ovum from the mother and a haploid spermatozoon from the father. The zygote then goes through many mitotic divisions to develop into an adult, with half of the genetic material in each cell from each parent. The adult, male or female, produces haploid gametes by meiotic cell division to repeat the life cycle once again.
The development of a zygote into an adult and the maintenance of adult cells and tissues requires many thousands of different gene products. All of these gene products are encoded in the genome and inherited from mother and father. The gene, a length of DNA coding for a particular gene product, is the fundamental unit of inheritance. [Are gene products always proteins?1] The genes are distributed among the chromosomes that compose the genome, and every gene can be pinpointed to a specific location called the locus (plural: loci) on a specific chromosome. [Can all physical traits of an organism be mapped to a single locus?2]
The human genome is split into 24 different chromosomes: 22 of these are autosomes (non-sex chromosomes) and 2 are sex chromosomes (or allosomes, X and Y). Each human has 23 pairs of chromosomes (22 autosomes and a sex chromosome), for a total of 46 chromosomes. One chromosome of each pair is from the mother and one is from the father. The two nonidentical copies of a chromosome are called homologous chromosomes. Although these two copies look the same when examined at the crudest level under a microscope, and although they contain the same genes, the copies of the genes in the two homologous chromosomes may differ in their DNA sequence. Different versions of a gene, called alleles, may carry out the gene’s function differently. Since a person carries two copies of every gene, one on each homologous chromosome, a person could potentially carry two different alleles. Individuals carrying different alleles of a gene will often have traits that allow the inheritance of alleles to be followed. [Is it possible for there to be more than two different alleles of a specific gene?3]
• Which one of the following is true if an individual has two different alleles at a given locus?4
A) The individual has two phenotypes, e.g., one brown eye and one blue.
B) There are two alleles in one place on one particular chromosome.
C) Two siblings have different appearances.
D) There is a different allele on each of the two members of a homologous pair.
Genotype vs. Phenotype
The genotype is the DNA sequence of the alleles a person carries. A person carrying two different alleles at a given locus is called a heterozygote, while an individual carrying two identical alleles is called a homozygote. The expression of alleles often is different in heterozygotes and homozygotes.
The phenotype is the physical expression of the genotype. For example, the phenotype of a gene involved in hair color may be brown or blond. Since there are many different kinds of alleles, there are different ways these alleles can be expressed in the phenotype. If an allele is the one expressed in the phenotype, regardless of what the second allele carried is, the expressed allele is referred to as dominant. An allele that is not expressed in the heterozygous state is referred to as recessive. For example, consider a heterozygous organism in which one allele encodes the functional version of an enzyme, while the second allele encodes an inactive version of that enzyme. Upon observation, it is noted that the organism’s enzymes are all functional; then the functional-enzyme allele is dominant and the inactive-enzyme allele is recessive. Since recessive alleles are not expressed in heterozygotes, it is not always possible to tell the genotype of an individual based solely on the phenotype. [Can a haploid organism like an adult fungus have recessive alleles?5]
There are certain conventions used in denoting genotypes in genetics that are useful to know. The alleles of a gene are usually denoted by letters. For example, for a gene called “curly,” a dominant allele may be denoted by the capital letter C and a recessive allele may be denoted by the lower case letter c. A heterozygote is referred to as Cc, while homozygotes would be either CC or cc. More complex situations require more complex conventions, but most questions probably only involve two alleles at a locus. [If the dominant allele for curly (C) results in curly hair and the recessive allele (c) causes straight hair, what are the phenotypes of CC, Cc and cc individuals?6]
8.2 MEIOSIS
Mitotic cell division produces two daughter cells that are identical to the parent. However, the production of haploid cells such as gametes from a diploid cell requires a type of cell division that reduces the number of copies of each chromosome from two to one; this method of cell division is called meiosis. In males, meiosis occurs in the testes with haploid spermatozoa as the end result; in females, meiosis in the ovaries produces ova. (Note: This is not always the case, and while meiosis begins in the ovaries, it is completed only after fertilization; see Chapter 14 for a further discussion on oogenesis.) Specialized cells termed spermatogonia in males and oogonia in females undergo meiosis. Spermatogenesis and oogenesis share the same basic features of meiosis but differ in many of the specific features of gamete production. Meiosis itself will be discussed in this chapter, while the specifics of spermatogenesis and oogenesis will be discussed in Chapter 14.
Mitosis and meiosis are similar in many respects. Mitosis and meiosis are both preceded by one round of replication of the genome (S phase), leaving a diploid cell with four copies of the genome (Figure 4). The different phases in cell division are referred to by the same names (prophase, metaphase, anaphase, and telophase) in both meiosis and mitosis and are mechanistically very similar. The primary difference between meiosis and mitosis is that replication of the genome is followed by one round of cell division in mitosis and two rounds of cell division in meiosis, meiosis I and meiosis II (Figure 5). Another important difference is that in meiosis, recombination occurs between homologous chromosomes.
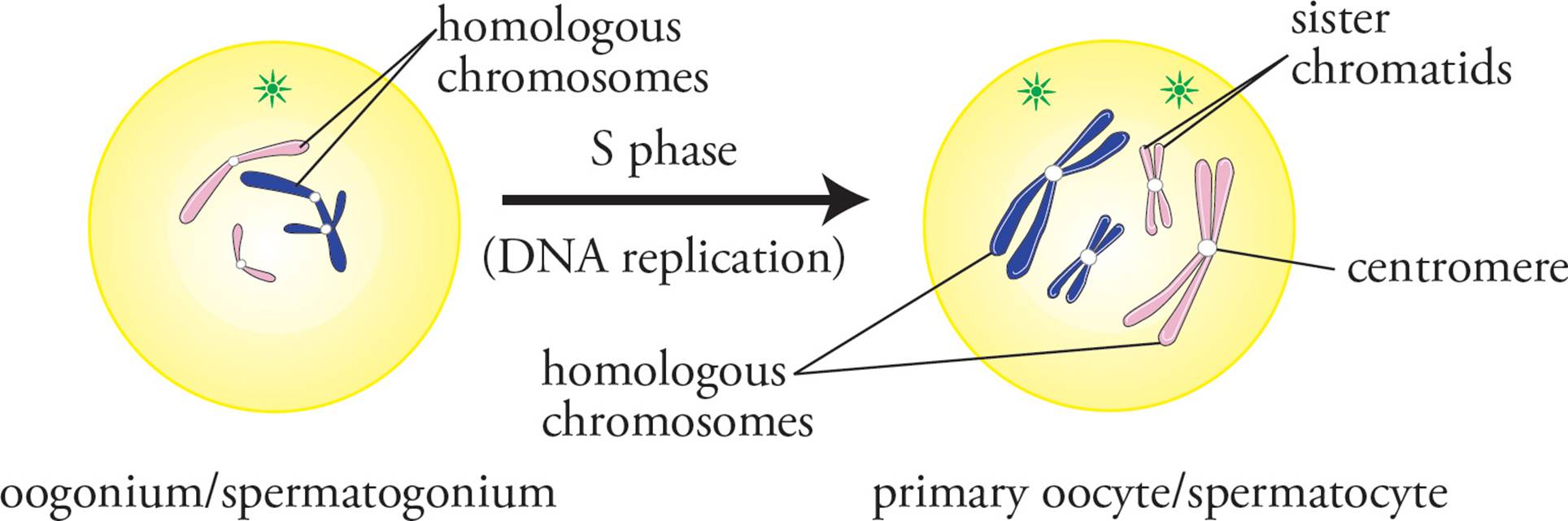
Figure 4 S-Phase
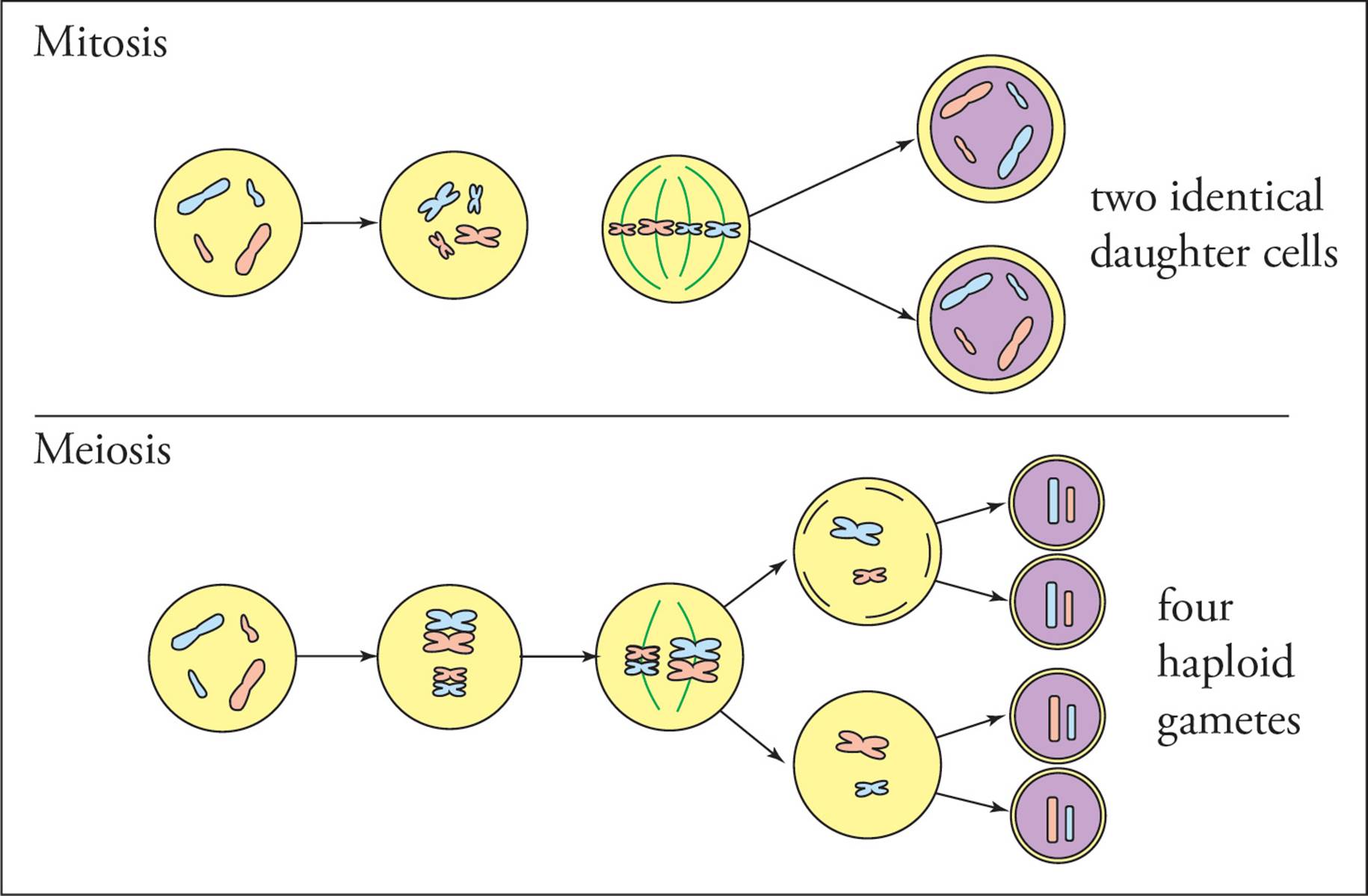
Figure 5 Mitosis vs. Meiosis
The first step in meiosis is prophase I (Figure 6). To depict meiosis, we will use a hypothetical model organism with a diploid genome with two different (nonhomologous) chromosomes (Figures 6–10).
• How many chromosomes are present in a cell from this organism during prophase I of meiosis?7
As in mitotic prophase, chromosomes condense in meiotic prophase I, and then the nuclear envelope breaks down. Unlike mitosis, however, homologous chromosomes pair with each other during meiotic prophase I in synapsis. Homologous chromosomes align themselves very precisely with each other in synapsis, with the two copies of each gene on two different chromosomes brought closely together. The paired homologous chromosomes are called a bivalent or tetrad.
When the DNA is aligned properly, it can then be cut precisely at the same location on homologous chromosomes. Genes are then swapped between the pair, and the chromosomes are religated (Figure 6). This process is known as crossing over or recombination (Figure 7). Due to the extreme complexity of crossing over, meiotic prophase takes the most time in meiosis, days sometimes. Recombination during meiosis is an important source of genetic variation during sexual reproduction.
• Does crossing over change the number of genes on a chromosome?8
• Does recombination create combinations of alleles on a chromosome that are not found in the parent?9
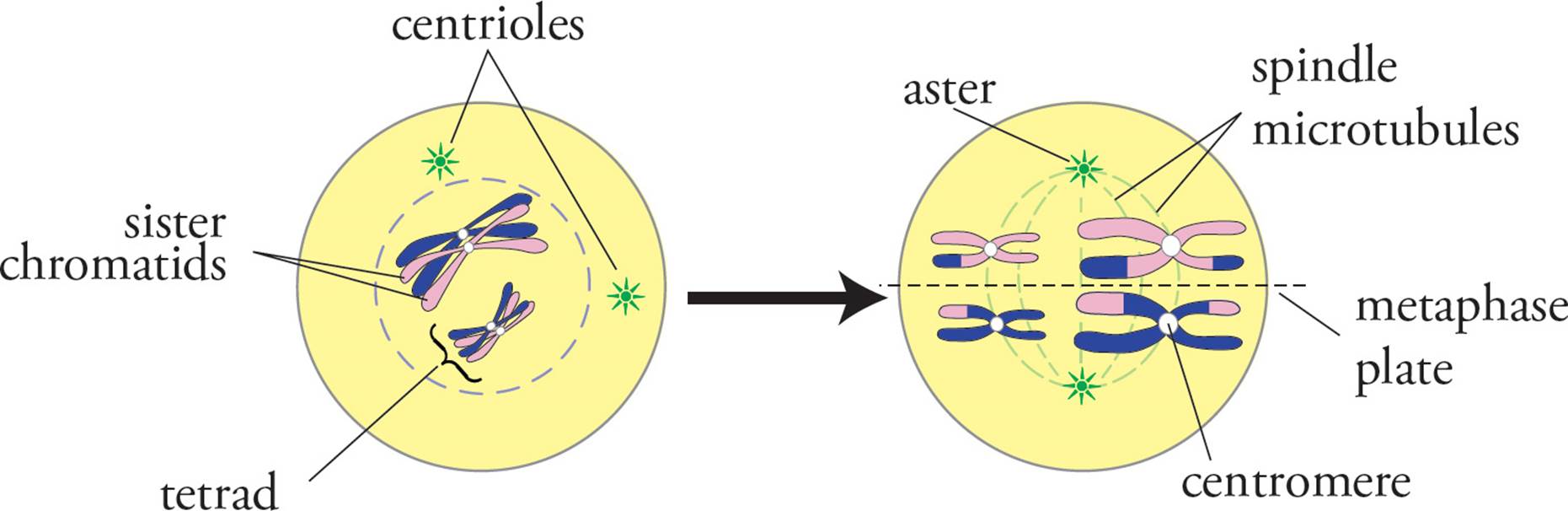
Figure 6 Prophase I and Metaphase I
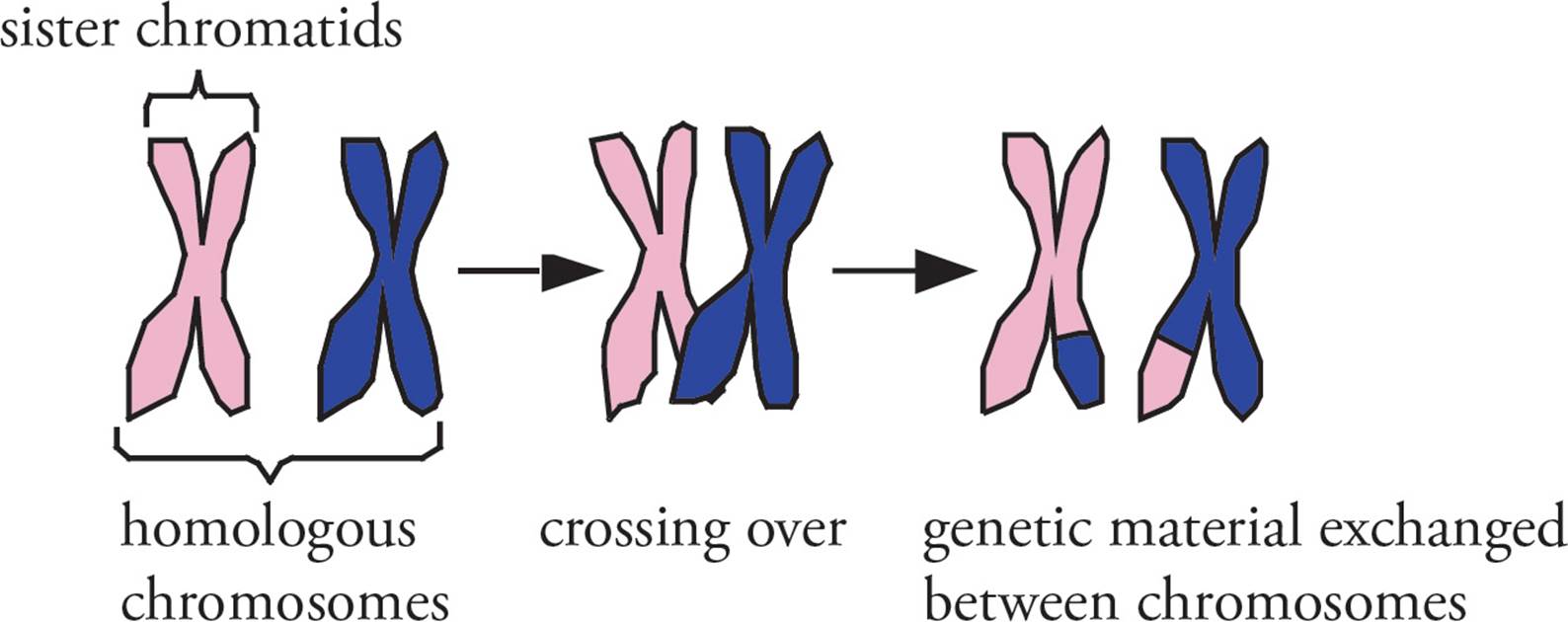
Figure 7 Crossing Over (Recombination)
Since precision is crucial in chromosome swapping, formation of the tetrad is highly regulated. Synapsis is mediated by a protein structure called the synaptonemal complex (SC). This structure starts to form early in meiotic prophase I. First, proteins named SYCP2 and SYCP3 attach to each of the two homologous chromatin structures that are to be paired (Figure 8). This makes up the lateral elements of the SC. The lateral regions then align and attach via a central region (made of SYCP1 and many other proteins). Both the lateral and central regions together form the SC, and essentially work like a zipper to connect homologous chromosomes.
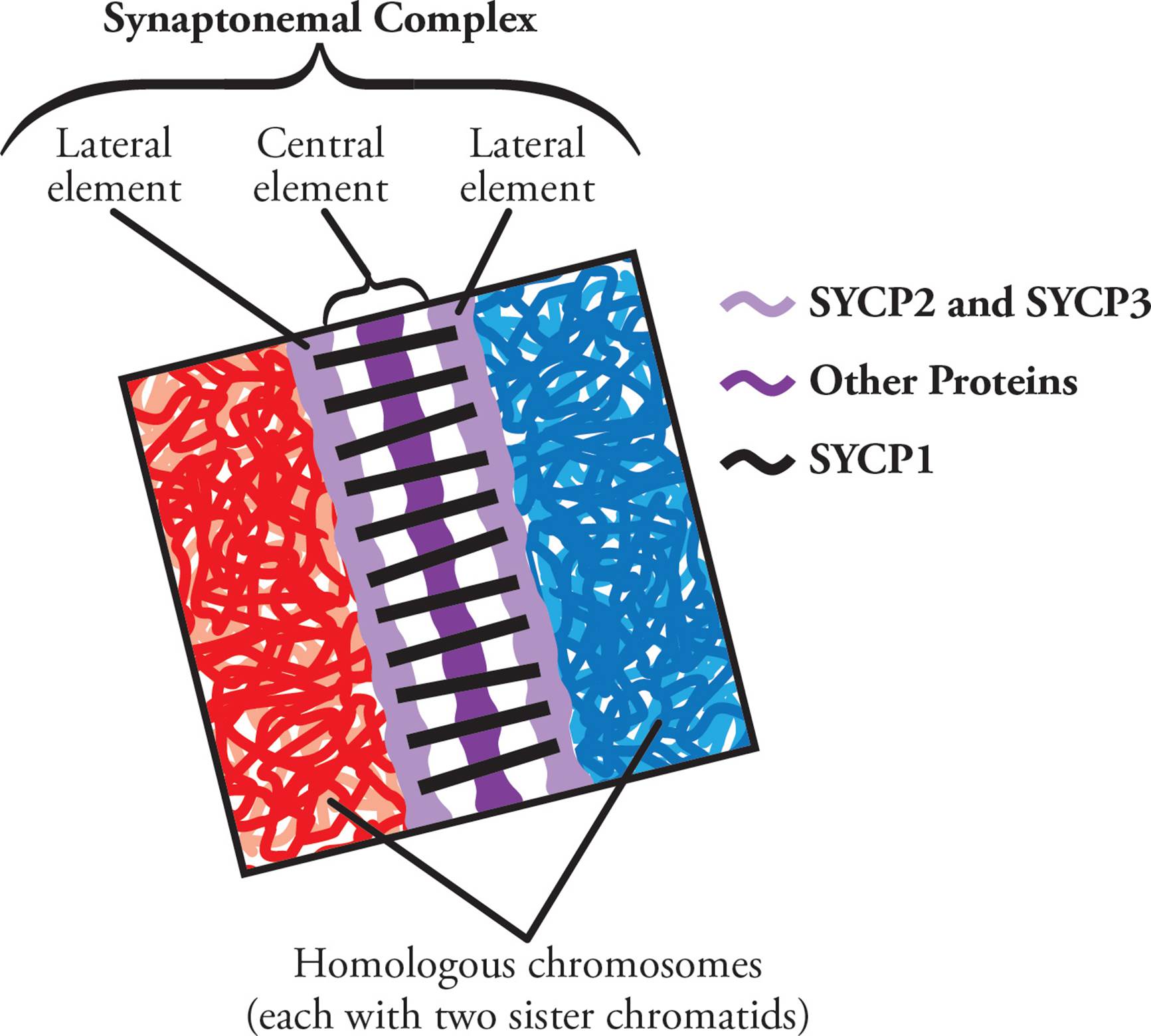
Figure 8 The Synaptonemal Complex
While no physical connection has yet been shown between the synaptonemal complex and recombination machinery, it’s been demonstrated that SC formation and recombination are interdependent. Both happen around the same time of meiosis, and work on mice with defective synaptonemal complex formation or recombination shows that these two processes rely on one another. When synaptonemal complex formation is inhibited, recombination is disturbed, and vice versa.
After prophase I is metaphase I. In meiotic metaphase I, alignment along the metaphase plate occurs, as in mitosis. The difference is that in meiotic metaphase I, the tetrads are aligned at the center of the cell (the metaphase plate), whereas in mitosis, sister chromatids are aligned on the metaphase plate. In anaphase I, homologous chromosomes separate, and sister chromatids remain together (Figure 9). The cell then divides into two cells during telophase I(Figure 10). It is important to note that at this point the cells are considered to be haploid. Each cell has a single set of chromosomes. The chromosomes, however, are still replicated (still exist as a pair of sister chromatids). The whole point to the second set of meiotic divisions is to separate the sister chromatids so that each cell has a single set of unreplicated chromosomes.
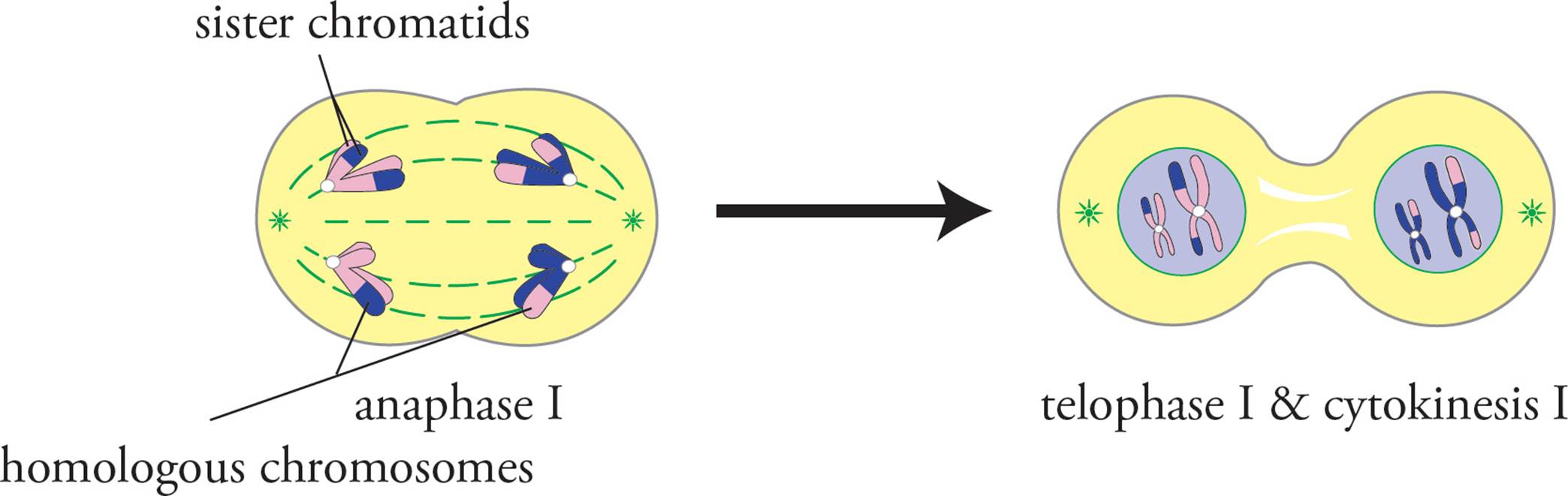
Figure 9 Anaphase I, Telophase I, and Cytokinesis I
In some species, meiosis II begins immediately after telophase I, while in other species, there is a period of time before meiosis II begins. In either case, there is no further replication of the DNA before the second set of divisions. The movements of the chromosomes during meiosis II are identical to the movements in mitosis, with the sole difference being that in meiosis II there is a haploid number of chromosomes, while in mitosis there is a diploid number. The sister chromatids are separated during anaphase II, and after telophase II is complete, four haploid cells have been produced from a single diploid parent cell (Figure 10).

Figure 10 Meiosis II
• When homologous chromosomes separate, do all paternal and maternal chromosomes stay together in the daughter cells?10
• Are the sister chromatids that separate during meiotic anaphase II identical in their DNA sequence?11
• Which of the following occur in meiosis but NOT in mitosis?12
I. Separation of sister chromatids on microtubules
II. Pairing of homologous chromosomes
III. Recombination between sister chromatids
A) I only
B) II only
C) I and II
D) II and III
• If cells are blocked in meiotic metaphase II and prevented from moving on in meiosis, which one of the following will be prevented?13
A) Crossing over
B) Separation of homologous chromosomes
C) Separation of sister chromatids
D) Breakdown of the nuclear envelope
Nondisjunction
Sometimes during meiosis I homologous chromosomes fail to separate, and sometimes during meiosis II sister chromatids fail to separate. Such a failure of chromosomes to separate correctly during meiosis is called nondisjunction. [A gamete normally contains how many copies of each chromosome?14 If two homologous chromosomes of chromosome #12 fail to separate during meiosis I, how many copies of chromosome #12 will the resulting gametes have?15] Gametes resulting from nondisjunction will have two copies or no copies of a given chromosome. Such a gamete can fuse with a normal gamete to create a zygote with either three copies of a chromosome (trisomy) or one copy of a chromosome (monosomy).
The genetic defect caused when an entire chromosome is either added or removed is usually so great that a zygote with either trisomy or monosomy cannot develop into a normal individual. There are examples in which nondisjunction is not lethal in humans, although it results in significant developmental abnormalities. Trisomy of chromosome #21 results in Down syndrome, with intellectual disability and abnormal growth. Nondisjunction of the sex chromosomes is also generally not lethal during development. Individuals who have only one X chromosome and no Y, for example, have Turner syndrome, with external female appearance but underdeveloped ovaries and sterility. Individuals with nondisjunction of the sex chromosomes will develop to have male appearance if they have at least one Y, no matter how many X chromosomes are present, and will have female genitalia if only X chromosomes are present. Most will be sterile, however, and many will suffer intellectual disability. [In an individual with Down syndrome, are the defects in development caused by an absence of genetic information?16 If not, why does trisomy of this chromosome or other chromosomes have such dramatic effects?17]
8.3 MENDELIAN GENETICS
Gregor Mendel described the statistical behavior of the inheritance of traits in pea plants long before the nature of DNA and chromosomes was known. Unlike Mendel, however, we are now familiar with the molecular basis of genetics in meiosis and genes, and the laws of genetics that Mendel formulated can now be presented with insight based on this knowledge. Although Mendelian genetics generally only involves the simplest patterns of inheritance, it forms the foundation for understanding more complicated situations.
Mendel observed that traits were governed by pairs of hereditary material (alleles). The first of Mendel’s laws, the law of segregation, states that the two alleles of an individual are separated and passed on to the next generation singly. [At what stage during meiosis are different alleles of a gene separated?18] Mendel’s second law, the law of independent assortment, states that the alleles of one gene will separate into gametes independently of alleles for another gene. We will illustrate these principles using the garden pea plant, but the principles apply equally well to humans.
A trait that can be studied in the pea plant is the color of the pea. We can call G the allele for green color, while g is the allele for yellow pea color. Mating between plants, a cross, is used as a tool in genetics to discern genotypes by looking at the phenotypes of progeny from a cross. A pure-breeding strain of yellow or green peas consistently yields progeny of the same color when mated within the strain. For example, if mating yellow plants with yellow plants always produces yellow progeny, yellow is a pure-breeding [Can anything be deduced about the genotype of the pure-breeding strain of yellow peas?19 If a pure-breeding yellow and pure-breeding green strain are crossed, and all of the progeny are green, what does this indicate about the expression of the yellow and green alleles?20] Let’s assume that G is the dominant allele of the color gene, and g is the recessive allele. [Is it possible to deduce the genotype of a pea plant at the color gene if it is green?21] If a green plant is encountered, a testcross can be performed to deduce the genotype of the plant. A testcross is when one individual is crossed to another individual that has a homozygous (or pure-breeding) recessive genotype. The presence of all recessive alleles in one parent allows alleles from the other parent to be displayed phenotypically. The progeny of a testcross are called the F1 generation. [If a green plant is testcrossed with a pure-breeding yellow strain, and some of the F1 generation are yellow while others are green, what is the genotype of the original green plant?22] The results of a testcross are dependent on statistics and follow Mendel’s laws.
The principle of segregation can be illustrated with the color gene described above for the pea. If a pea is heterozygous Gg, its gametes will contain either the G allele or the g allele, but never both. [If a gamete contained both G and g, what occurred during meiosis?23] The probability that a gamete in the heterozygote will contain one allele or the other is 50%, completely random. [Would the principle of segregation apply to a gene on the X chromosome in a woman?24] To illustrate the law of independent assortment, we need to introduce a second gene, one that controls the shape of the pea. W is the dominant allele, resulting in wrinkled peas, while w is the recessive allele, resulting in smooth peas in homozygous ww plants. According to the law of independent assortment, the genes for the color of peas and the shape of peas are passed from one generation to another independently. [If the color gene and the shape gene are right next to each other on a chromosome, will they display independent assortment?25] The nature of the shape gene in a given gamete does not depend on and is not influenced by the color gene, if independent assortment is true. [If an individual is heterozygous at the color gene, Gg, and heterozygous at the shape gene, Ww, what are the chances that a gamete containing the G allele will also contain the W allele?26]
The Punnett Square
It is possible to predict the results of a cross between two individuals using the laws of segregation and independent assortment. Determining the result can be complex, however, so a visual tool called the Punnett square is often employed to make the process simpler. Let’s use a simple square first, with only one trait involved (Figure 11); we will then tackle a more complicated problem with two different traits (Figure 12).
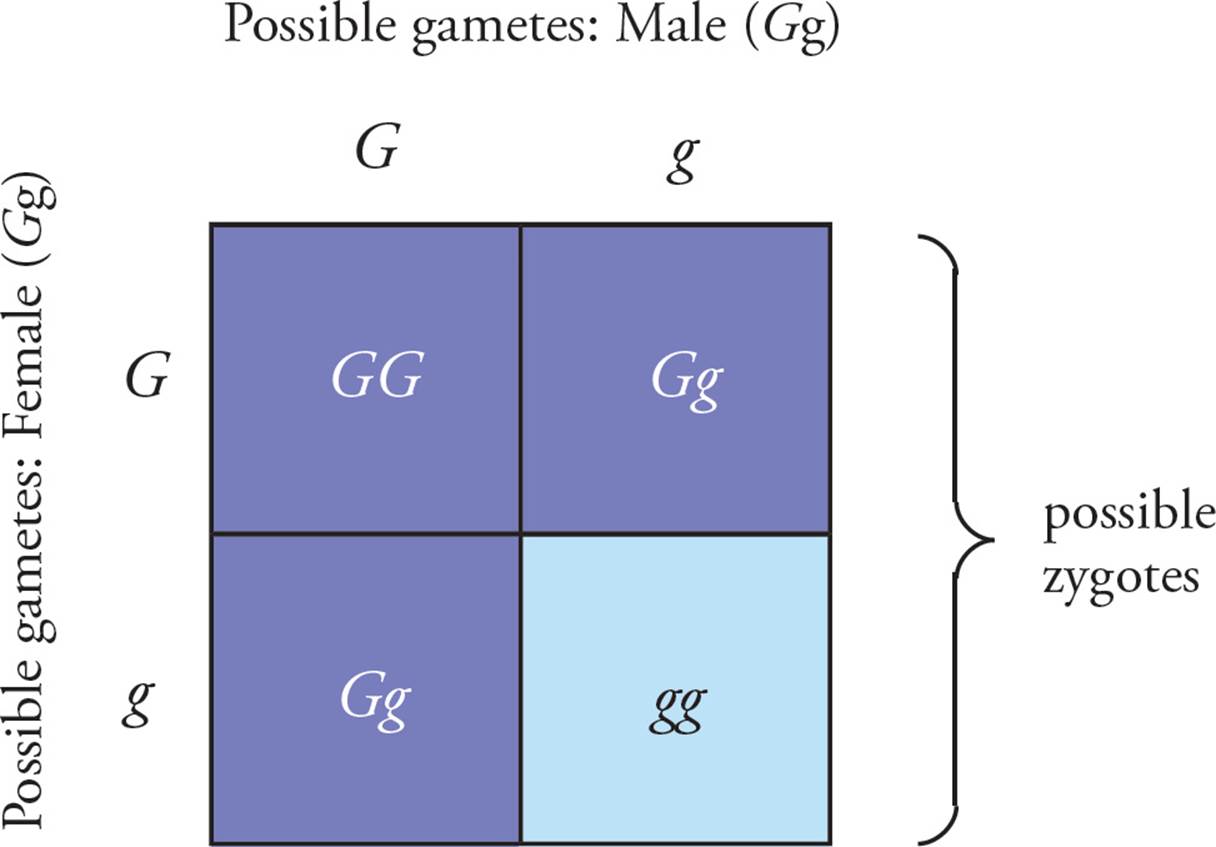
Figure 11 A Punnett Square Involving One Gene
In Figure 11, a Punnett square depicts a cross between two pea plants that are heterozygous for the color gene, with G the dominant green allele and g the recessive yellow allele. To draw a Punnett square, the following steps are involved:
Step 1: Determine the gametes that are possible from each parent in the cross.
Step 2: Draw a square with the possible gametes from each parent on two sides.
Step 3: Fill in the square with the zygote genotypes that would result from each possible combination of gamete.
Step 4: Determine the phenotype of each genotype.
Step 5: Find the probability of each genotype and each phenotype.
• In the situation shown in Figure 11, which one of the following will be true?27
A) 25% of the offspring will be green, and 75% will be yellow.
B) 50% of the offspring will be green, and 50% will be yellow.
C) 75% of the offspring will be green, and 25% will be yellow.
D) 100% of the offspring will be green.
A more complicated Punnett square is needed to look at two traits during a cross. In Figure 12, a cross is performed between Plant 1, heterozygous at the color gene (Gg), and Plant 2, also heterozygous at the color gene. Plant 1 is also homozygous for the dominant allele of the shape gene (wrinkled peas) while Plant 2 is homozygous for the recessive allele (smooth). [What are the phenotypes of the plants being crossed?28] The same steps are followed to construct the Punnett square in Figure 12 as the one in Figure 11. First, determine the possible gametes for each pea plant being crossed. (In this case, there are really two possible gamete types from each parent, so the box could be simplified to have only two gametes on a side). Then, determine the possible combinations of gametes that could join to form zygotes and the phenotypes and frequencies of the F1 generation. [What percentage of the F1 generation will have smooth peas?29 What percentage of peas will be green and wrinkled?30 Yellow and wrinkled?31 The cross depicted in Figure 12 was performed and produced 77 green wrinkled plants and 20 yellow wrinkled plants; why do these results not agree exactly with the ratios predicted in the Punnett square?32] Independent assortment and the principle of segregation are assumptions built into this Punnett square.
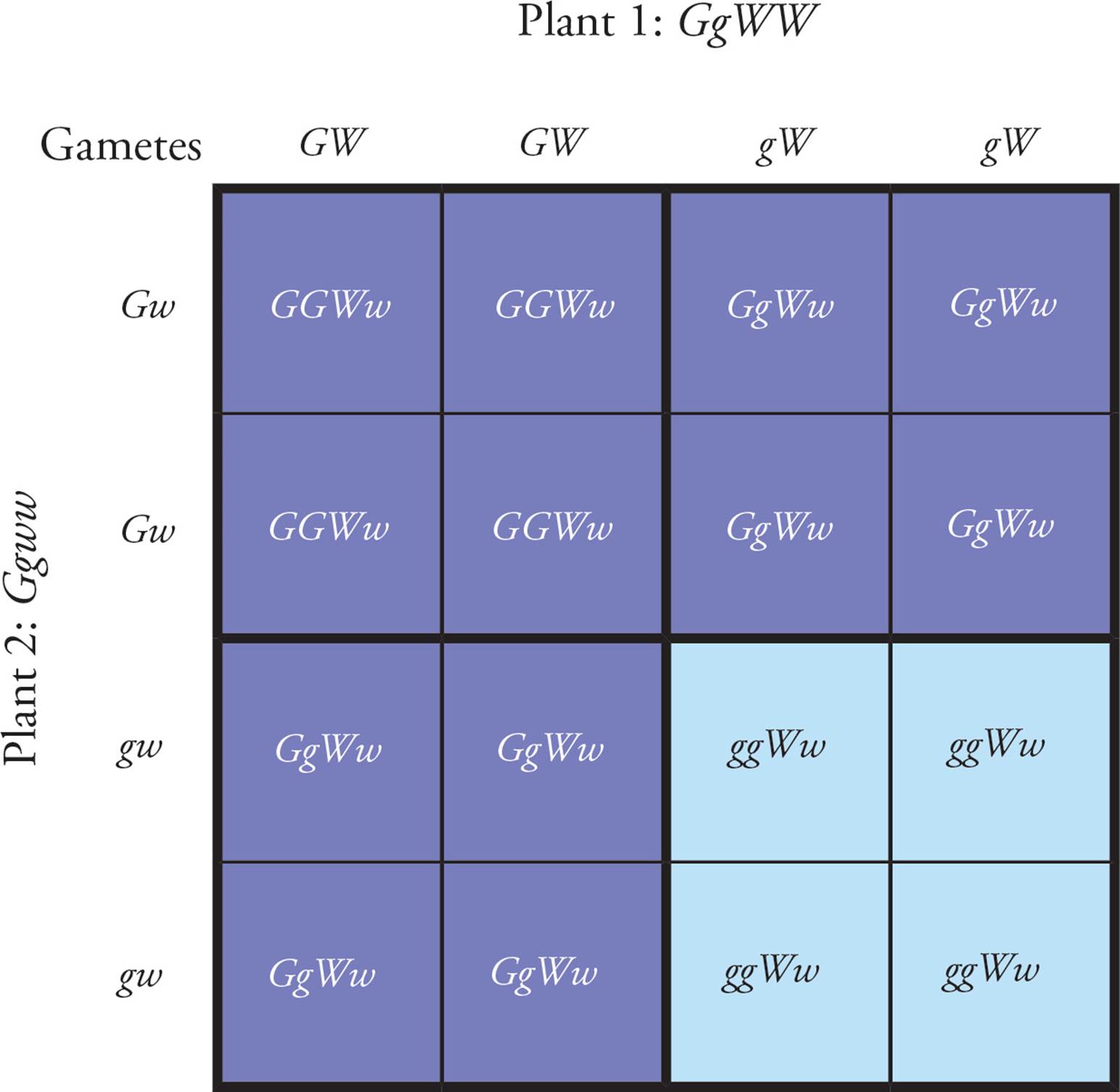
Figure 12 A Punnett Square Depicting a Cross with Two Traits Involved
• In the cross depicted in Figure 12, how does the shape gene affect inheritance of the alleles for the color gene?33
A) The percentage of green peas is increased by the shape gene.
B) The shape gene has no effect on the inheritance of the alleles for the color gene.
C) The percentage of green peas is decreased by the shape gene.
D) The shape gene prevents segregation of the alleles for the color gene.
• If a green wrinkled plant from the F1 generation in Figure 12 is crossed with a pure-breeding yellow smooth pea plant, what phenotypes are possible?34
• If any yellow smooth progeny are observed in this testcross, what does this indicate about the genotype of the F1 plant?35
The Rules of Probability
Punnett squares are only one way to determine the probability of an outcome in a cross. Another way involves using statistical rules called the rule of multiplication and the rule of addition. The rule of multiplication states that the probability of both of two independent events happening can be found by multiplying the odds of either event alone. For example, if the probability of being struck by lightning is 1 in a million (10–6) and the probability of winning the lottery is 10–7, then the probability of both happening is the product: 10–6 × 10–7 = 10–13.
The rule of addition can be used to calculate the chances of either of two events happening. The chance of either A or B happening is equal to the probability of A added to the probability of B, minus the probability of A and B occurring together. For example, the chance of either getting hit by lightning or winning the lottery is 10–6 + 10–7 = 1.1 × 10–6. (Note: The product of 10–6 and 10–7 is so small that it can be neglected from the equation.) These rules can be a shortcut to using a Punnett square in some problems.
• A man that is homozygous for eye color, bb, is married to a woman who is heterozygous at the same gene: Bb. What are the chances that a child will have the Bb genotype and be a boy?36
Other Biostatistical Methods
Biologists use many other statistical methods in genetics, and many other areas. The chi-square test is used to compare observed and expected data, and t-tests are used to compare two data sets. Data is often summarized using mean, median and mode. Finally, standard deviation and standard error give an indication of how spread out a dataset is. Each of these is reviewed in Appendix II.
8.4 EXTENDING MENDELIAN GENETICS
Mendel first started his work using mice, but abandoned rodents (either because of the mess involved, or because of the questionable ethics of a monk studying breeding schemes). In picking pea plants instead, he made a fortuitous decision. The traits he chose to study are (for the most part) controlled by a single gene with two alleles each. These two alleles have one completely dominant to the other, and therefore a simple relationship between genotype and phenotype. Although Mendel’s peas all displayed very simple patterns of inheritance, the inheritance of traits is often more complicated.
Incomplete Dominance
Some alleles of genes display neither dominant nor recessive patterns of expression. If the phenotype of a heterozygote is a blended mix of both alleles, this is called incomplete dominance, and the alleles for that trait are given different, upper-case letters. For example, if a gene for flower color has two incompletely dominant alleles, R could be used to indicate the allele for red color and W to indicate the allele for white color. [If a gene for flower color has two alleles, R (red) and r (white), and R is dominant while r is recessive, what is the phenotype of Rr heterozygotes?37 If R and W display incomplete dominance, what is the phenotype of RW heterozygotes?38 How many phenotypes are possible if R and W display incomplete dominance?39]
Codominance
Codominance is a slightly different situation, in which two alleles are both expressed but are not blended. For example, the alleles of the gene for ABO blood group antigens that are found on the surface of red blood cells display codominance. Each of the alleles is expressed on red blood cells, regardless of the second allele in the cell. There are three alleles for the ABO blood group antigens: I A, I B, and i. The alleles IA and IB are codominant and will be expressed regardless of the second allele, while i is recessive to both IA and IB. The alleles IA and IB cause type A or type B antigens to be expressed, while i does not cause antigen expression.
• What is the phenotype of an individual heterozygous for the IA and I B alleles?40
• What is the phenotype of an individual heterozygous for I B and i?41
• If a woman heterozygous for type A blood marries a man who is heterozygous for type B blood, what are the possible genotypes (and blood types) of their children?42
The other main antigen used in blood typing is the Rh (rhesus) factor. The expression of this antigen follows a classically dominant pattern; RhDRhD and RhDRhd (also seen as RR and Rr) genotypes lead to the expression of this protein on the surface of the red cell (Rh positive), and the RhdRhd (or rr) genotype leads to the absence of the protein (Rh negative).
Although Mendel’s peas all displayed very simple patterns of inheritance, there are often many complications in the inheritance of traits. For example:
|
Pleiotropism: |
A gene is said to have pleiotropic effects if its expression alters many different, seemingly unrelated aspects of the organism’s total phenotype. For example, a mutation in a gene may cause altered development of heart, bone, and inner ears. |
|
Polygenism: |
Complex traits that are influenced by many different genes are called polygenic. These traits tend to display a range of phenotypes in a continuous distribution. For example, height is polygenic and is influenced by genes for growth factors, receptors, hormones, bone deposition, muscle development, energy utilization, and so on. As a consequence, there is a wide range of normal heights for adult humans; we are not just “tall” and “short,” like Mendel’s peas. Skin color and mouse fur color are additional examples of mammalian polygenic traits. |
|
Penetrance: |
Penetrance describes the likelihood that a person with a given genotype will express the expected phenotype. While many traits are completely penetrant (all individuals with a given allele or mutation display the phenotype), there is a spectrum of options: alleles or mutations can also have high, incomplete, or low penetrance. The root cause of penetrance depends on the allele. Some have age-related penetrance, where the phenotype is displayed more frequently in mutation-carrying individuals as they age. The penetrance of other alleles depends on environmental and lifestyle modifiers. For example, women who carry a certain mutation that increases their risk of breast cancer display variable rates of breast cancer, depending on their diet, if they smoke, if they have had children and breast fed, etc. Finally, many alleles have genetic modifiers that affect penetrance; since several human traits are polygenic, alleles at different loci can affect penetrance. |
|
Epistasis: |
This refers to a situation where expression of alleles for one gene is dependent on a different gene. For example, a gene for curly hair cannot be expressed if a different gene causes baldness. |
|
Recessive Lethal Alleles: |
Some mutant alleles can cause death of an organism when present in a homozygous manner. These are called recessive lethal alleles, and they typically code for essential gene products. In diploid organisms, these alleles can be studied by maintaining heterozygous stocks, which are then mated together to form a homozygous recessive offspring. Embryonic development studies can shed light on when this organism dies and possibly why. Studying recessive lethal alleles in haploid organisms is much harder. Here, a conditional system is usually used where the allele is normal (or permissive) under certain conditions, allowing survival of the organism. The mutant allele can be induced under different conditions (such as a different temperature), to study effects of the allele. |
• 100 people are homozygous for an allele that is implicated in cancer, but only 20 develop cancer. What are potential explanations for why only some people express a gene out of a broader population with the same genotype?43
• In one strain of mouse, homozygotes for an allele of a gene develop heart defects, while in another strain of mouse, homozygotes with the same allele develop normally. Heterozygotes develop normally in both strains. What is the most likely explanation for the difference between the two strains?44
A) The allele is recessive.
B) The development of the heart defect is influenced by more than one locus.
C) The allele has pleiotropic effects on development.
D) The allele is codominant.
The Sex Chromosomes
Early in the twentieth century it was observed that women have twenty-three pairs of chromosomes that are homologous, while men have only twenty-two pairs of chromosomes that match in appearance. The two chromosomes in men that did not match each other were termed the X and the Y chromosomes because of their appearance during mitosis (Figure 13). Males have an X and a Y, while females have two X chromosomes. The presence of a Y chromosome in humans (genotype XY) is a key factor in the determination of the sex of an embryo, and subsequent development into a male. The absence of a Y (genotype XX) results in a female as the default developmental pathway. During meiosis, females generate gametes that contain an X chromosome; males generate gametes with either an X or a Y chromosome, meaning that it is the male gamete that determines the gender of an embryo (Figure 14).
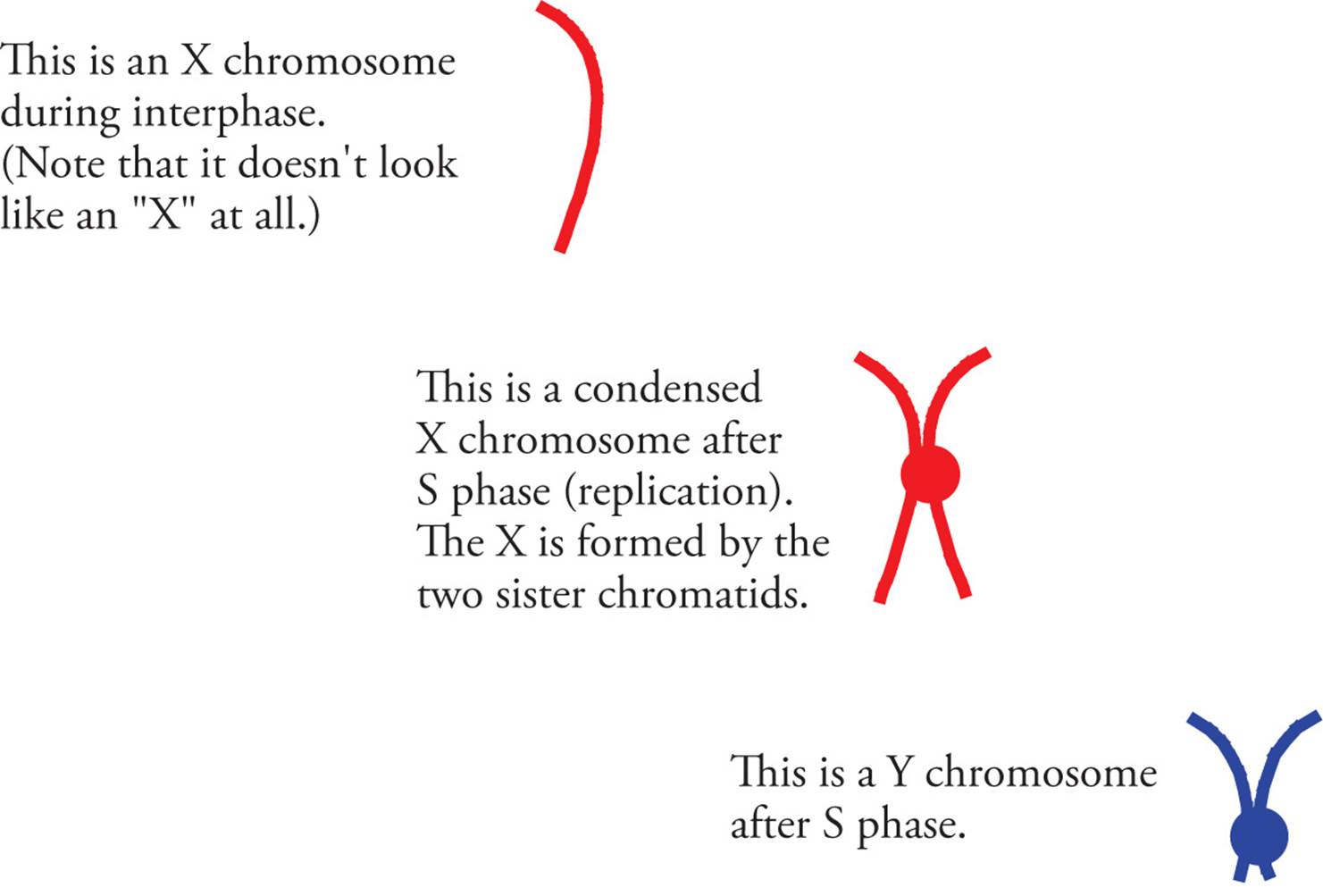
Figure 13 The Sex Chromosomes
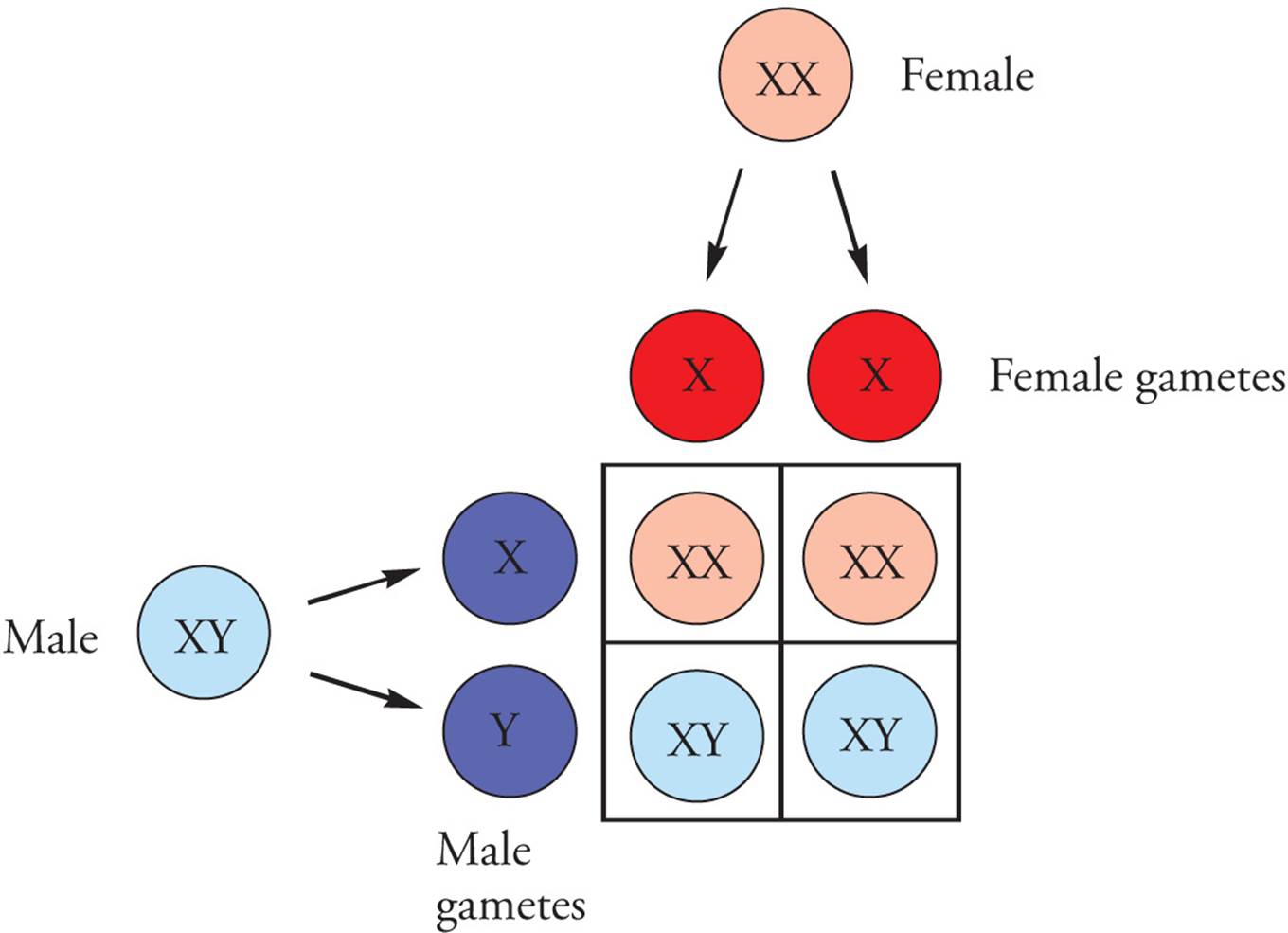
Figure 14 Determination of the Zygote’s Sexual Genotype
The sex chromosomes also play a key role in the inheritance of other traits that are not directly involved in sexual development. Much of what has been discussed about inheritance was dependent on the assumption that there are two copies of every chromosome and therefore two copies of every gene in each cell. This is true for genes found on every pair of chromosomes except for one pair: the sex chromosomes. Genes that lie on the X chromosome will be present in two copies in females but only in one copy in males. [What pattern of expression will a recessive allele on the X chromosome display in males?45] Traits that are determined by genes on the X or Y chromosome are called sex-linked traits because of their unique patterns of expression and inheritance. The inheritance of traits coded by genes on sex-chromosomes will be covered in Section 8.6.
8.5 LINKAGE
The traits that Mendel studied and based the law of independent assortment on were located on separate chromosomes. Genes that are located on the same chromosome may not display independent assortment, however. The failure of genes to display independent assortment is called linkage.
• If eye color is controlled by a gene on chromosome #11 and the hair color locus is located on chromosome #14, do these genes assort independently?46
• If the portion of chromosome #14 containing the hair color gene is translocated onto chromosome #11, will these genes still assort independently?47
If genes are located very close to each other on the same chromosome, then they will probably not be inherited independently of each other. Let’s illustrate this with a pea gene for height and two alleles of the height gene, tall (T) and short (t), with the T allele dominant and the t allele recessive. If the height gene and the color gene are very near each other on the same chromosome, then the alleles of these genes on a specific chromosome will probably assort together into gametes during meiosis (Figure 15). This limits the possible combinations of the alleles in the gametes.
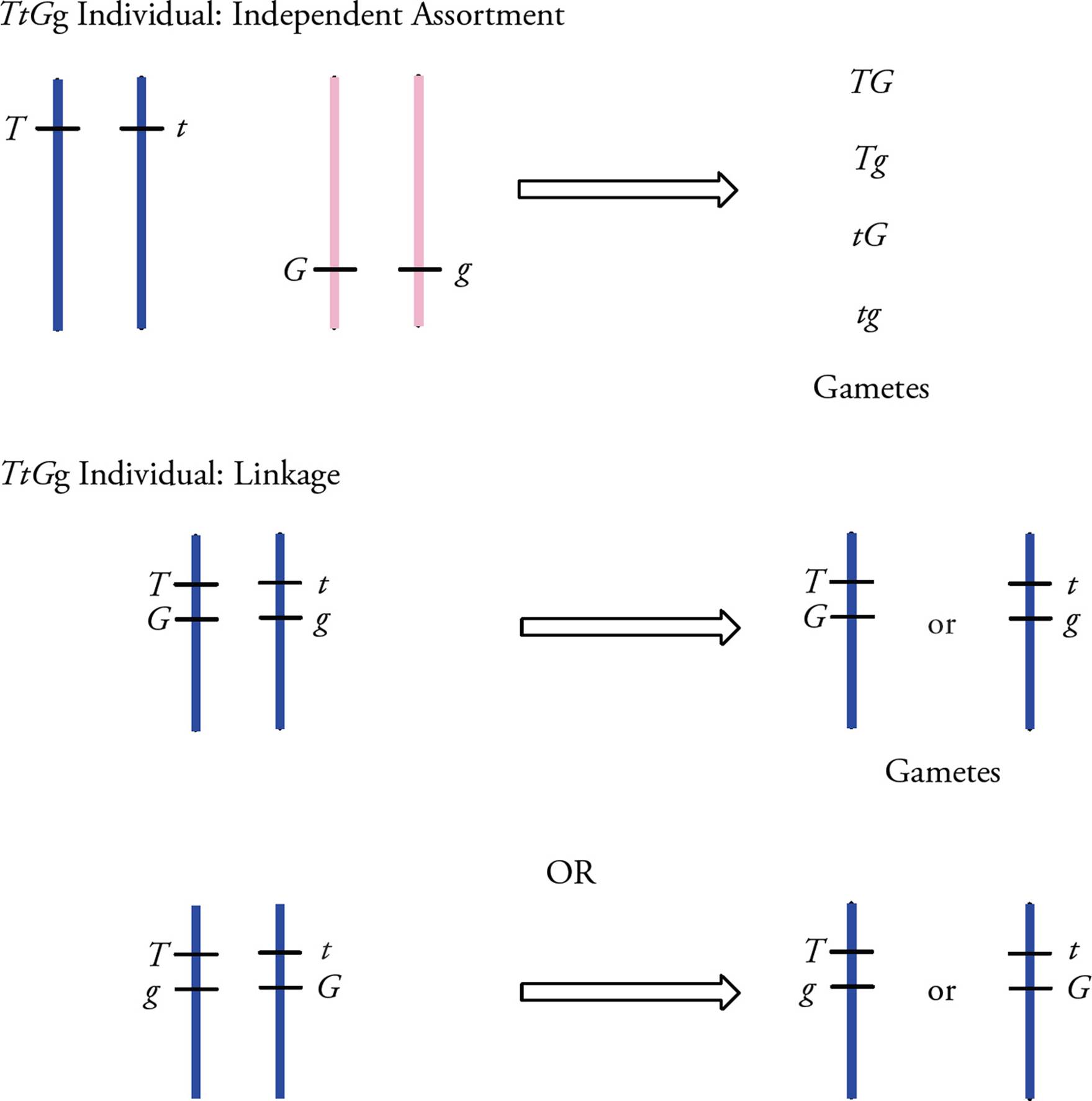
Figure 15 Linkage of Alleles during Meiosis
• If the color gene and the height gene display linkage, is it possible to predict the possible gametes of a TTgg individual?48 of a TtGg individual?49
To know how alleles that display linkage assort during meiosis, it may be necessary to know which alleles were on a chromosome together. As seen in Figure 15, there are two possible ways the height and color genes could be linked. The dominant alleles of two different genes can be linked together on the same chromosome (TG), the recessive alleles of two different genes can be linked (tg), or one dominant and one recessive allele can be linked (Tg and tG).
With genes that are found on the same chromosome, the design of a Punnett square is slightly different. The possible gametes are limited since they cannot assort independently. Consider a cross between a homozygous ttgg pea plant and a double-heterozygous plant with both dominant alleles on one chromosome and both recessive alleles located together on another chromosome. They can only make a limited number of different gametes, not the four possible combinations of alleles that would be found if the genes were on different chromosomes. A Punnett square will help to illustrate linkage in this example (Figure 16).
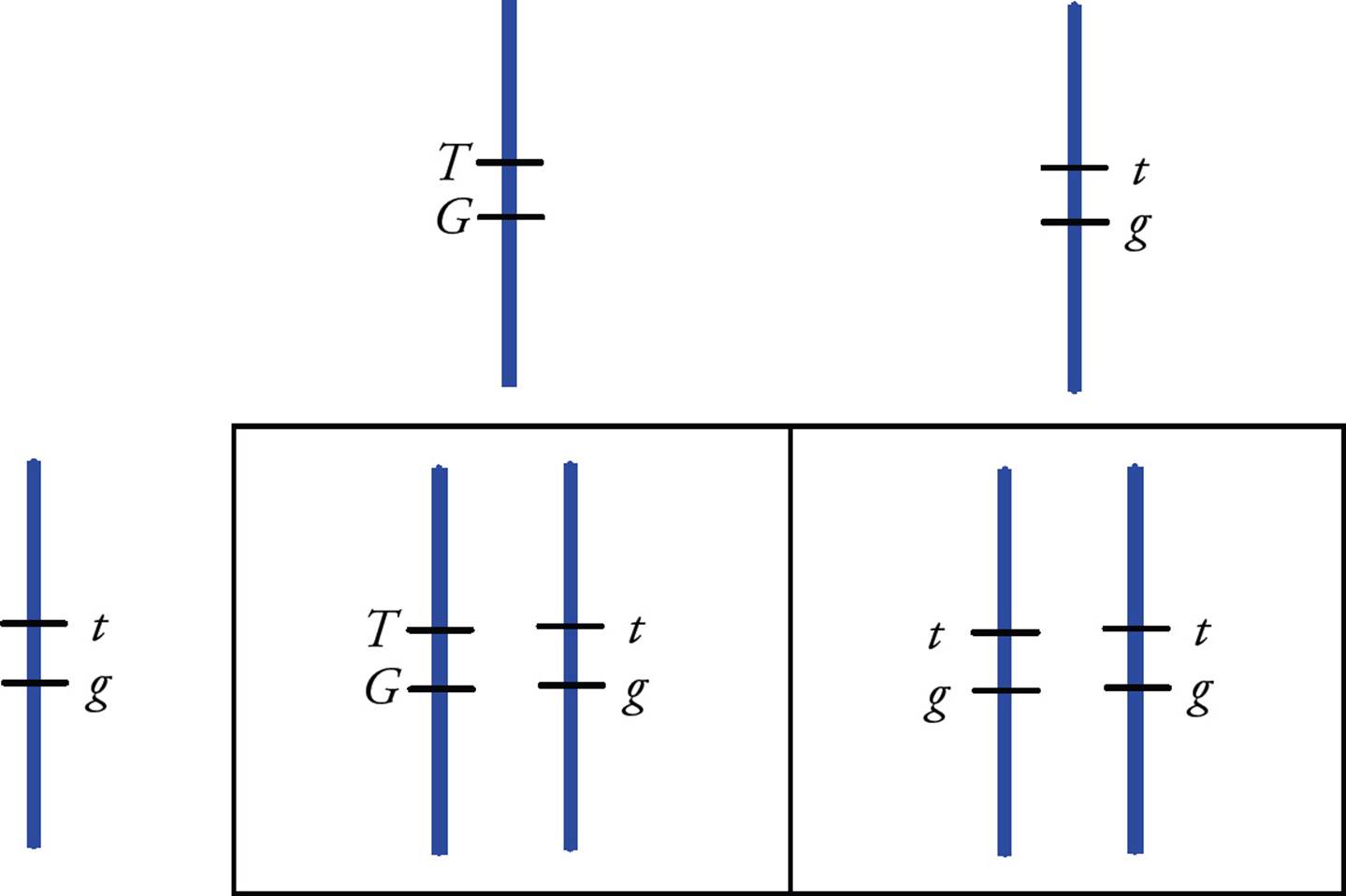
Figure 16 Assortment of Linked Genes
• What are the phenotypes of the F1 progeny in the cross in Figure 16?50
• If a tall green pea from the F1 progeny is crossed with a pure-breeding short yellow plant, what phenotypes will be observed and in what ratios?51
• If height and color genes were not linked, what ratios of phenotypes would be observed in a cross between a TtGg and a ttgg individual?52
• Assume all of the characteristics already introduced for the height, color and shape pea genes that have been used as examples. The height and color genes are located near each other on the same chromosome and display complete linkage but the shape gene is located on a different chromosome. If an individual with a TtGgWw genotype and the T and g alleles on the same chromosome is crossed with a ttGgWw individual, what result will be observed?53
A) All tall peas will be wrinkled.
B) All wrinkled peas will be tall.
C) All yellow peas will be tall.
D) All tall peas will be yellow.
Linkage and Recombination
Linkage is the exception to the law of independent assortment. When genes are located on the same chromosome, they will display linkage and will not assort independently. Meiotic recombination provides the exception to linkage. During the formation of gametes, meiotic recombination between homologous chromosomes can separate alleles that were located on the same chromosome. In the example in Figure 17, three genes are located on the same chromosome. Prior to recombination, ABC were found on one chromosome and abc were found on the homologous chromosome. [What combinations of alleles will be found in gametes in the absence of recombination?54] Recombination produces new combinations of alleles not found in the parent and also allows genes located on the same chromosome to assort independently.
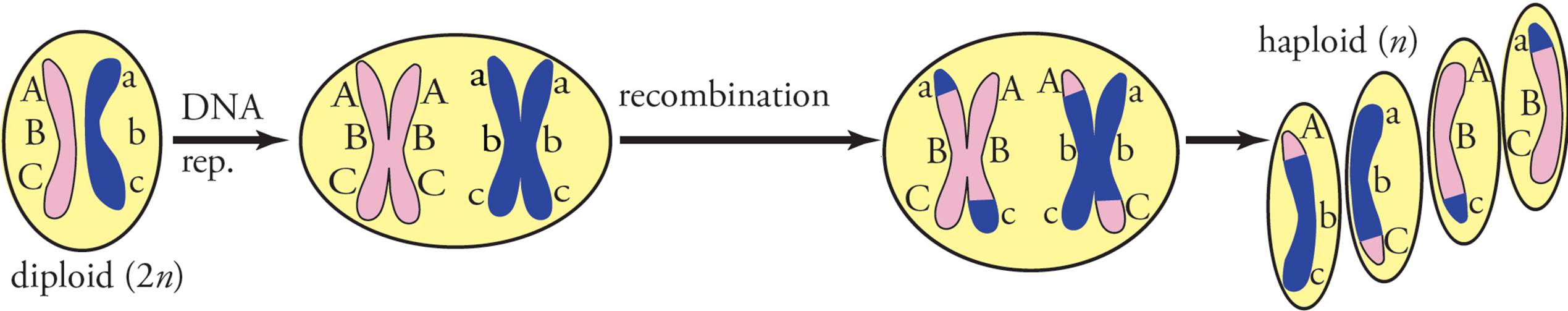
Figure 17 Recombination—Another Look
The example of the height and color genes in pea plants will help to illustrate linkage and the effects of recombination on patterns of inheritance. As before, the height and color genes are located on the same chromosome. There are two alleles of the height gene, dominant T (tall) and recessive t (short) and two alleles of the color gene, dominant G (green) and recessive g (yellow). The following cross is performed: A pure-breeding tall green plant is crossed with a pure-breeding short yellow plant. [What phenotypes are predicted in this cross if linkage is complete?55] A pea plant from this cross is then self-pollinated (crossed with itself) to produce an F2 generation. [If linkage is complete, what genotypes and phenotypes will be observed in the F2 generation?56 If the genes assort completely randomly, what genotypes and phenotypes will be observed in the F2 generation?57] The F2 generation in this cross was observed to have the following plants: 30 tall green plants, 9 short yellow plants, 2 tall yellow plants, and 1 short green plant. [Which of these phenotypes are recombinant phenotypes?58] Often in a cross involving genes on the same chromosome, the result will be intermediate between independent assortment and complete linkage. The reason for this is that recombination occurs between the genes during meiosis of some of the gametes but not all of the gametes. [If it is known that two genes are located on the same chromosome but during a cross they assort completely randomly, how can this be?59]
The frequency of recombination between two genes on a chromosome is proportional to the physical distance between the genes along the linear length of the DNA molecule. [Does recombination occur between genes more frequently if they are near each other or far apart?60] The farther apart two genes are on a chromosome, the more likely recombination will occur between the genes during meiosis. If the genes are located far enough apart, recombination will occur so frequently between the genes that they will no longer display linkage and will assort as independently as if they were on separate chromosomes. The frequency of recombination is given as the number of recombinant phenotypes resulting from a cross divided by the total number of progeny.
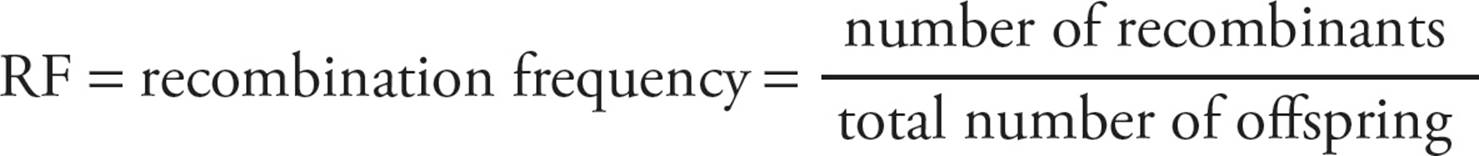
Since the frequency of recombination is proportional to the physical distance of genes from each other, it can be used as a tool to map genes in relation to each other on chromosomes.
Example: The height and color gene in pea plants are on the same chromosome as a third gene for big or small flowers. The alleles of flower size are a dominant B (big) and a recessive b (small). The color gene (G, green or g, yellow) is studied in relation to the flower size gene. In the first cross, pure-breeding homozygous BBGG plants are crossed with bbgg plants. [What is the phenotype of the F1 progeny?61 If a small flower green plant is observed in the F1 generation, was recombination responsible?62] An F1 progeny is then crossed with a bbgg plant and the following phenotypes observed: 44 big flower green plants, 40 small flower yellow plants, 8 big flower yellow plants, and 8 small flower green plants.
• Which of these are recombinant phenotypes?63
• What is the frequency of recombination between the genes?64
• What is the maximal frequency of recombination?65
• In another cross, the frequency of recombination between the flower size and height genes is examined and found to be 10 recombinant plants out of 100 progeny. Is the height gene or the color gene closer to the flower size gene?66
• If the recombination frequencies are 0.16 between the height and color genes, 0.10 between the height and flower size genes, and 0.26 between the flower size and color genes, what is the order of the genes on the chromosome?67
• Is it possible to map the distance between genes on the same chromosome even if they are so far apart that they assort independently?68
• Assume that hair color in humans is determined by a gene for which there are two alleles: B or brown, which is dominant, and b or blond, which is recessive. The hair color gene is located on the same chromosome as another gene that determines the strength of bones, and the two genes are very close together. The alleles of the bone strength gene are S, the dominant sturdy bone allele, and s, the recessive fragile bone allele. Jose and Tonya have dark hair and sturdy bones. One of their children has brown hair and fragile bones. One grandparent of Jose and one grandparent of Tonya had fragile bones and blond hair, while the remaining grandparents were homozygous for brown hair and sturdy bones. Which of the following is/are true?69
I. The child of Jose and Tonya represents a recombinant phenotype.
II. All of the children of Jose and Tonya must have fragile bones.
III. Jose and Tonya may have other children with blond hair and fragile bones.
A) I only
B) II only
C) I and III only
D) II and III only
8.6 INHERITANCE PATTERNS AND PEDIGREES
There are six inheritance patterns that you should be familiar with: autosomal recessive, autosomal dominant, mitochondrial, Y-linked, X-linked recessive and X-linked dominant. In this section, each will be described and then a summary Table is presented.
Autosomal traits are caused by genetic variation on the autosomes (the 22 pairs of non-sex chromosomes in humans). These traits can be autosomal dominant (in which case a single copy of the allele will confer the trait or disease phenotype) or autosomal recessive (in which case two copies of the allele are required for the affected phenotype). Both tend to affect males and females equally; in other words, there is no sex bias for these traits.
There is a small, haploid DNA genome inside the mitochondria and humans inherit this genome from their mothers. This is because the sperm contributes only nuclear chromosomes to the zygote; the ovum contributes nuclear chromosomes and the rest of the cellular material including the organelles. There are some traits that are inherited via the mitochondrial genome, although these mitochondrial traits are rare. Luckily, they are fairly easy to spot because affected females have all affected offspring (sons and daughters). Affected individuals must have an affected mother, and affected males cannot have any affected offspring. An individual cannot inherit mitochondrial traits from their father. Mitochondrial traits (like Y-linked traits and X-linked traits in human males) are an example of hemizygosity; the individual only has one copy of the chromosome in a diploid organism. Because of this, there is only one allele to keep track of for each individual. Genes encoded by the mitochondrial genome are usually given the prefix mt (for example, mt-Atp6 is encoded in the mitochondrial genome and codes for a subunit of the ATP synthase). When working with inheritance patterns though, it is best to define the allele letters you are going to use and then use one letter per individual. For example, you could assign “a” as a normal individual and “A” as an affected individual. The assignment here is arbitrary since one allele is not dominant to the other (they are mutually exclusive since humans only have one mitochondrial genome). The key is to be consistent.
Traits that are determined by genes located on the X or Y chromosome are called sex-linked traits and display unusual patterns of inheritance. Traits encoded by genes on the Y chromosome (Y-linked traits) would only be passed from male parents to male children. [Would it be possible for a father to pass a Y-linked trait to female children?70 Can males be carriers of recessive Y-linked traits without expressing them?71] Y-linked traits are quite rare, because the Y chromosome is small and contains a relatively small number of genes. Many of the genes on the Y-chromosome function in sex determination.
X-linked traits are observed quite frequently and can be X-linked recessive or X-linked dominant. There are several well-studied examples of X-linked recessive traits that are common in the human population; hemophilia is an example. Women are often carriers of X-linked recessive alleles but only express recessive X-linked traits when they are homozygous. Men are hemizygous for X-linked traits; they have only one copy of genes on the X chromosome. As a result, males always express recessive X-linked alleles. [From which parent do males receive X-linked traits?72] These traits tend to affect males more than females.
Red-green colorblindness, an X-linked trait, is caused by a defect in a visual pigment gene on the X chromosome. The allele that is responsible for colorblindness is a pigment gene that does not produce functional protein. [Is the colorblindness allele recessive or dominant?73] The colorblindness allele, like many recessive traits carried in the population, is not expressed in heterozygotes. Colorblindness is unusual in women but fairly common in men. Females have two copies of the gene, so will not express the trait if they are heterozygotes, while males have only one X chromosome and so will always express the allele whenever they receive it. [A man is colorblind, and his wife is homozygous normal for genes encoding visual pigment proteins. What will be the phenotypes and genotypes of sons and of daughters of this couple?74]
X-linked dominant traits are harder to identify. A female will display an X-linked dominant phenotype if she has one or two copies of the allele on her X chromosomes. A male will express the phenotype if he inherited the affected allele from his mother. While these traits still tend to affect males more than females, this trend is less obvious than for X-linked recessive traits.
Table 1 on the following page summarizes the six inheritance patterns you should be familiar with, and lists some strategies you can use to distinguish between them.
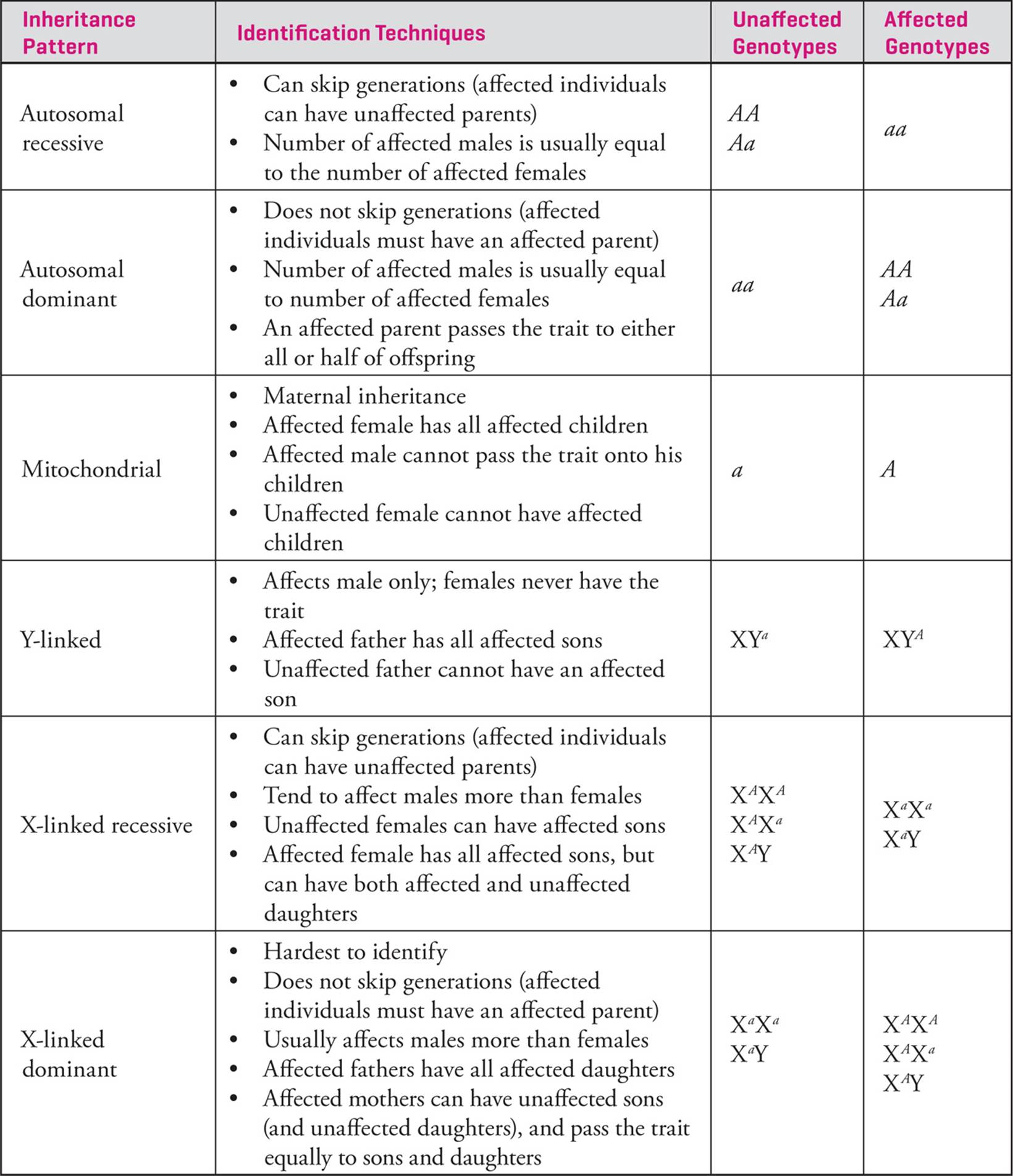
Table 1 Summary of Inheritance Patterns
• Two mouse genes located on the X chromosome are being studied. The alleles of the genes are:
Fuzzy hair: F, dominant (normal hair) and f, recessive (fuzzy hair)
Extra toes: E, dominant (extra toes), and e, recessive (normal toes)
A female with normal hair and extra toes is crossed with a male with normal hair and extra toes. The progeny have the following phenotypes:
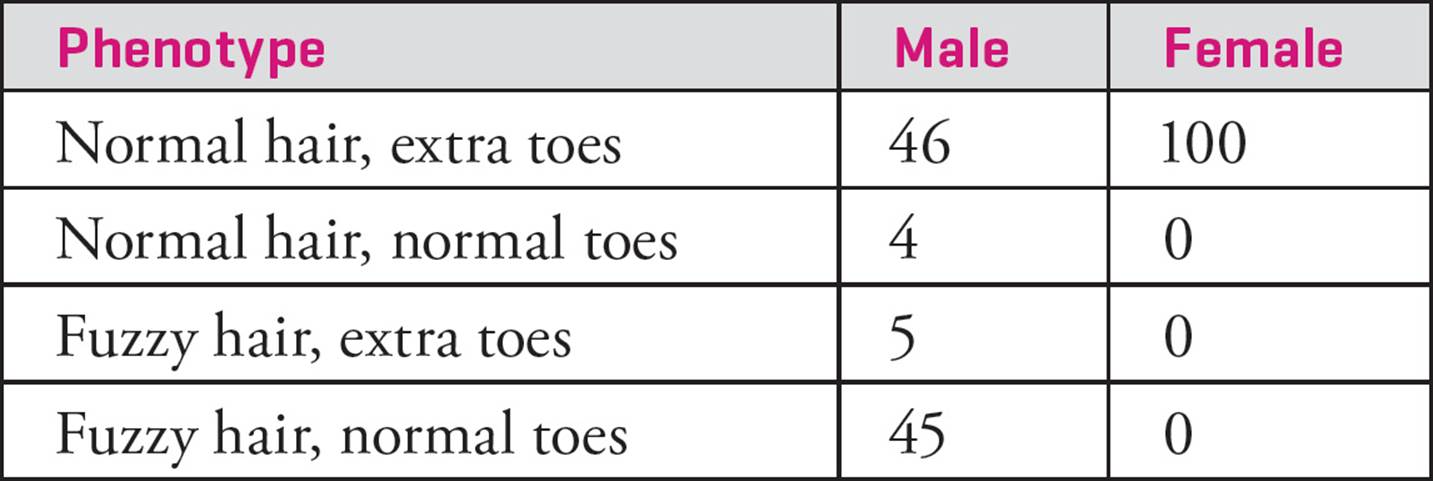
Which one of the following is true concerning this experiment?75
A) Males have a higher rate of recombination than females do.
B) In the absence of recombination, all males would have normal hair and extra toes.
C) The rate of recombination on the X chromosome is the same in males and females.
D) Both males and females have recombinant genotypes, but only males have recombinant phenotypes.
Often it is not possible to perform controlled genetic crosses to ascertain the nature of inheritance of a trait, particularly when people are involved. In these cases, families can be studied to determine the pattern of inheritance. Researchers organize the information learned from families into pedigrees, which are charts depicting inheritance of a trait (Figure 18). By studying the pedigree of families, researchers can determine the pattern of inheritance of a gene, whether it is linked to other genes, and whether an individual is likely to pass on a trait to their offspring. Pedigrees follow certain conventions in how they are drawn:
1) Males are represented by squares and females by circles.
2) A cross (mating) between a male and female is represented by a horizontal line connecting them.
3) Offspring from a cross are connected to their parents by a vertical line, and to each other by a horizontal line with vertical branches for each sibling.
4) Offspring of uknown gender (unborn children) are represented by a diamond shape.
5) Individuals afflicted with a trait being studied are shaded in; unaffected or normal individuals are not shaded in.
Many pedigrees make a common assumption: individuals mating into the family (i.e., individuals for which you have no information on their parents or grandparents) are assumed to be homozygous normal unless their phenotype tells you differently. The basis of this assumption is that the traits being studied are usually relatively rare in the human population and therefore it is most likely that a non-family member is homozygous for the wild type allele.
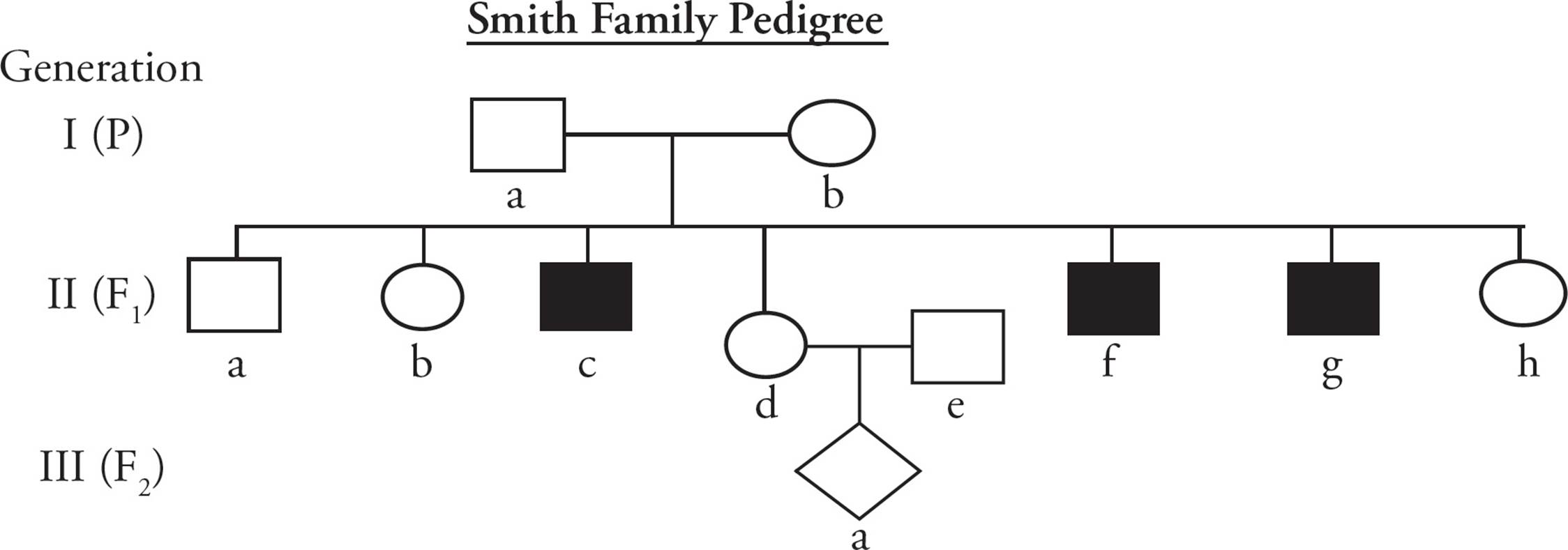
Figure 18 A Pedigree
Once drawn, a pedigree can be analyzed as follows:
Step 1: Is the allele that causes the trait dominant or is it recessive? Recessive traits commonly skip generations (affected individuals can have unaffected parents), but dominant traits do not (affected individuals must have at least one affected parent).
Step 2: Is the gene involved carried on a sex chromosome (sex-linked)? If so, there tends to be an unequal distribution of affected males (more) vs. affected females (fewer). If the numbers of affected males and females are approximately equal, the gene is most likely autosomal.
Step 3: If the disease is sex-linked, is it on the X or the Y chromosome? Diseases linked to the Y chromosome will show father-to-son transmission, while diseases linked to the X chromosome will not.
Step 4: Check for mitochondrial inheritance. Affected females will have all affected children, but affected males cannot pass the trait on.
Step 5: Figure out the genotypes and calculate the probabilities of inheritance where necessary. When writing genotypes for sex-linked traits, make sure to include the chromosomes (e.g., XAY, or XAXa, etc.). When writing genotypes for autosomal traits, make sure NOT to include the chromosomes (e.g., DD or Dd, etc.).
Step 6: If more than one trait is involved, go through Steps 1–5 for each.
• In the pedigree in Figure 18, the darkened squares represent individuals afflicted with a certain genetic disease. This disease is most likely caused by:76
A) a dominant allele.
B) an autosomal recessive allele.
C) an X-linked recessive allele.
D) a Y-linked allele.
• In the pedigree in Figure 18, what is the probability that IIIa will have the disease?77
A) If male, IIIa will have the disease.
B) Overall, there is a 1/8 chance that IIIa will have the disease.
C) Overall, there is a 1/4 chance that IIIa will have the disease.
D) IIIa will not have the disease.
Below there are example pedigrees for six modes of inheritance (X-linked recessive, X-linked dominant, autosomal recessive, autosomal dominant, mitochondrial, and Y-linked). For each pedigree, determine which mode of inheritance is displayed.
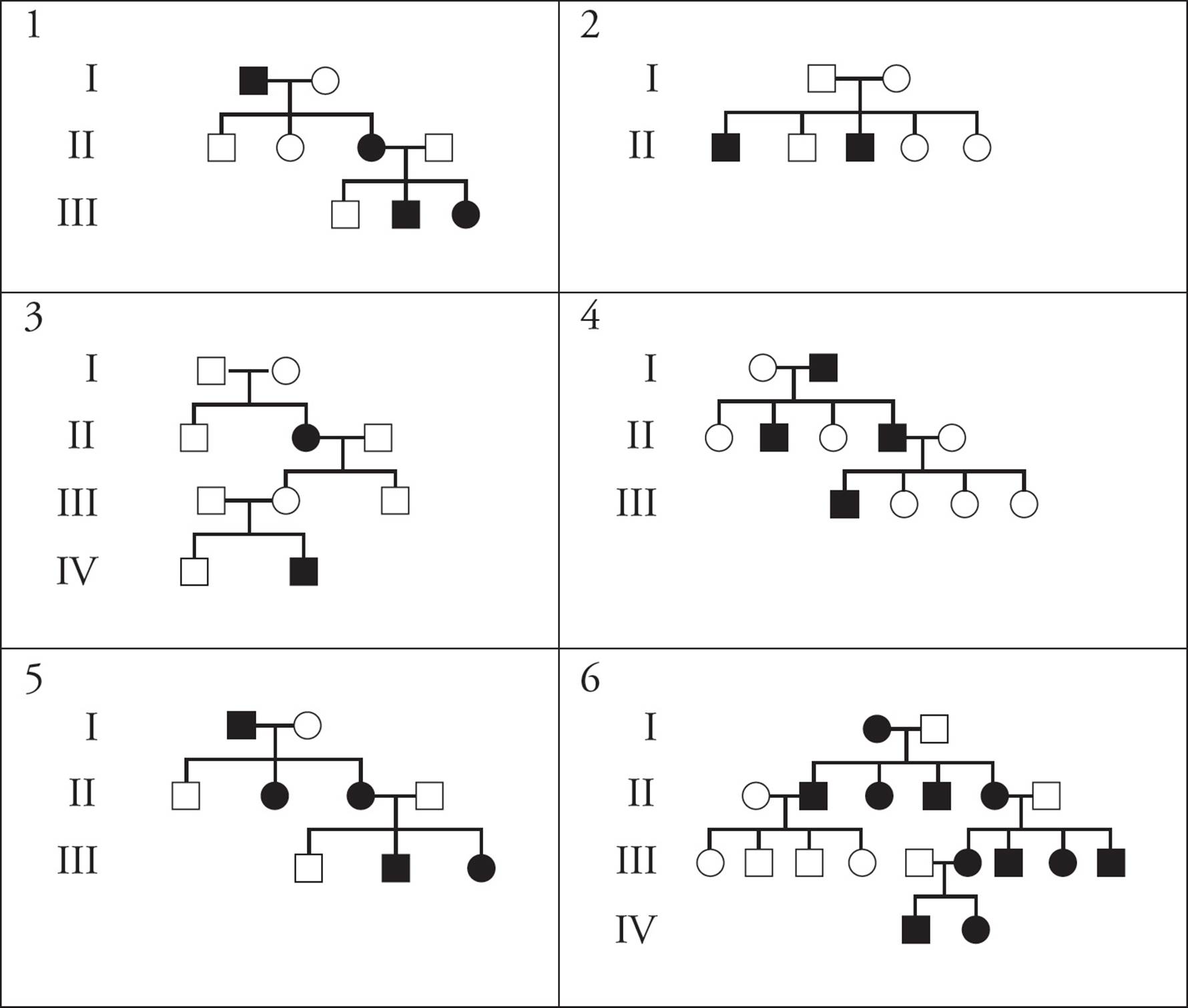
Answers
1) autosomal dominant
2) X-linked recessive
3) autosomal recessive
4) Y-linked
5) X-linked dominant
6) mitochondrial inheritance
Explanations
The easiest inheritance patterns to spot are mitochondrial (passed from mothers to all offspring) and Y-linked (passed from fathers to sons, and females are never affected). Let’s start by finding these two. Since Pedigree 6 shows a trait with maternal inheritance, this must be mitochondrial inheritance. The affected father in generation II does not pass the trait to any of his children, but the affected mothers in generations I, II and III pass the trait onto all their offspring. Pedigree 4 shows a trait with Y-linked inheritance. The trait is passed from father to all sons and does not affect females.
Next, Pedigrees 2 and 3 both show traits that skips generations. That is, there are individuals on the pedigree that are affected by the trait but who have unaffected parents. Therefore, these two pedigrees must be for recessive traits. One is autosomal and the other is X-linked.
Since the trait on Pedigree 2 affects males more than females, it is likely X-linked, and since the trait on Pedigree 3 affects males and females equally, it is probably autosomal. Let’s verify this by looking more closely at Pedigree 3. If this trait is X-linked recessive, then the affected female in generation II must have the genotype XaXa, and would have had to receive the allele for the trait from both her parents. However, for an X-linked recessive, the unaffected male in generation I would have the genotype XA Y, and would only have XA to donate to his daughter. Therefore this pedigree cannot represent an X-linked recessive trait; it must represent an autosomal recessive trait. The male in generation I must have the genotype Aa, the female in generation I must have the genotype Aa, and the affected female in generation II must have the genotype aa. The remaining pedigree, Pedigree 2, must be X-linked recessive.
Finally, Pedigrees 1 and 5 show traits that do not skip generations. That is, affected individuals have affected parents. These are pedigrees for dominant traits; one is X-linked and one is autosomal. The only difference between these two pedigrees is the middle daughter in the second generation; in Pedigree 1 she is unaffected and in Pedigree 5 she is affected. Let’s focus on her father (the male in generation I) since this is where she gets the allele for the trait. If the trait is X-linked dominant, the male in generation I would be XA Y; thus all females in generation II would inherit XA from their father, and all of them would be affected. Since the middle daughter in Pedigree I is not affected, Pedigree 1 must show autosomal dominance and the pedigree for the X-linked dominant trait must be Pedigree 5.
8.7 POPULATION GENETICS
Mendelian genetics describes the inheritance of traits in the progeny of specific individuals. For the purposes of large topics such as natural selection and evolution, however, the more relevant issue is not the inheritance of traits from individuals but in a whole population from one generation to another. Population genetics describes the inheritance of traits in populations over time. The word population has a specific meaning in this setting: a population consists of members of a species that mate and reproduce with each other. [If a group of sea turtles lives most of the year dispersed over a large area of ocean without contact with each other but congregate once a year to reproduce, is this group a population?78] To a population geneticist, each individual is merely a temporary carrier of the alleles in a population.
In population genetics, the units of genetic inheritance are alleles of genes, just as in Mendelian genetics. However, in population genetics alleles are examined across the entire population rather than in individuals. The sum total of all genetic information in a population is called the gene pool. [For an autosomal gene in a population of 2000 individuals, how many copies of the gene are present in the gene pool?79] The frequency of an allele in a population is a key variable used to describe the gene pool. [If there are 5000 hippos in a population, out of which there are 100 homozygotes of an autosomal allele h and 400 heterozygotes, what is the frequency of the h allele in the population?80If 20% of the population is heterozygous for an allele Q and 10% is homozygous, what will be the frequency of the allele in the population?81]
Hardy-Weinberg in Population Genetics
Population genetics does not simply describe the gene pool of a population but attempts to predict the gene pool of a population in the future. The Hardy-Weinberg law states that the frequencies of alleles in the gene pool of a population will not change over time, provided that a number of assumptions are true:
1) There is no mutation.
2) There is no migration.
3) There is no natural selection.
4) There is random mating.
5) The population is sufficiently large to prevent random drift in allele frequencies.
What Hardy-Weinberg means at the molecular level is that segregation of alleles, independent assortment, and recombination during meiosis can alter the combinations of alleles in gametes but cannot increase or decrease the frequency of an allele in the gametes of one individual or the gametes of the population as a whole.
• If 100 homozygous green pea plants and 100 homozygous yellow pea plants are crossed, 1000 green pea plants are produced. Does this mean that the yellow alleles disappeared from the population?82
• What is the frequency of the yellow allele in the gene pool of the progeny?83
• If the green peas from the F1 generation are allowed to mate randomly within the population, and there is no mutation, migration, natural selection, or random drift, what will be the frequency of the yellow allele in the population after four generations?84
• If two genes are closely linked on the same chromosome, will Hardy-Weinberg still apply to these genes?85
• According to Hardy-Weinberg, what will happen to the frequency of the yellow allele if predation occurs on yellow plants, but yellow plants attract bees more successfully?86
The Hardy-Weinberg law has also been translated into mathematical terms. Assuming that there are two alleles of a gene in a population, the letter p is used to represent the frequency of the dominant allele, and the letter q is used to represent the frequency of the recessive allele. Since there are only two alleles, the following fundamental equation must be true:
p + q = 1
Based on allele frequency, it is possible to calculate the proportion of genotypes in a population. Take a situation where the frequency of a dominant allele, G, equals p and the frequency of a recessive allele, g, equals q. If the equation above is squared on both sides, it becomes:
(p + q)2 = 1
p2 + 2pq + q2 = 1
where
p2 = the frequency of the GG genotype
2pq = the frequency of the Gg genotype
q2 = the frequency of the gg genotype
• If the frequency of the G allele is 0.25 in a population of 1000 mice, determine the number of individuals who are Gg heterozygotes if there is random mating but no migration, mutation, random drift, or natural selection.87
• If allele frequencies in a population are constant, and genotype frequencies can be calculated from allele frequencies, how will genotype frequencies vary over time?88
After one generation, a population will reach Hardy-Weinberg equilibrium, in which allele frequencies no longer change. Since allele frequencies do not change, and genotype frequencies can be calculated from allele frequencies, it follows that genotype frequencies also do not change over time. [If 100 green peas (GG) and 100 yellow peas (gg) are allowed to mate randomly, will the genotype frequencies in the next generation (F1) be the same?89 If not, why not?90 If the plants are allowed to mate randomly for another generation (F2), will the genotype frequencies in the F1 and F2 generations be the same?91]
Hardy-Weinberg in the Real World
Hardy-Weinberg requires a number of assumptions in order to be true. The assumptions, as presented earlier, are that in a population there is random mating and no mutation, migration, natural selection, or random drift. Thus, Hardy-Weinberg describes a highly idealized set of conditions required to prevent alleles from being added or removed from a population. In reality, it is not possible for a population to meet all of the conditions required by Hardy-Weinberg.
1) Mutation: Mutation is ineviTable in a population. Even if there are no chemical mutagens or radiation, inherent errors by DNA polymerase would over time cause mutations and introduce new alleles in a population.
2) Migration: If migration occurs, animals leaving or entering the population will carry alleles with them and disturb the Hardy-Weinberg equilibrium.
3) Natural Selection: For there to be no natural selection, there would have to be unlimited resources, no predation, no disease, and so on. This is not a set of conditions encountered in the real world.
4) Non-random Mating: If individuals pick their mates preferentially based on one or more traits, alleles that cause those traits will be passed on preferentially from one generation to another.
5) Random Drift: If a population becomes very small, it cannot contain as great a variety of alleles. In a very small population, random events can alter allele frequencies significantly and have a large influence on future generations.
8.8 EVOLUTION BY NATURAL SELECTION
At one time, life on Earth was generally viewed as static and unchanging, but we now know that this is not the case. Over the geologic span of Earth’s history, many species have arisen, changed over millions of years, given rise to new species, and died out. These changes in life on Earth are called evolution. Although he did not arrive at his theory alone, Charles Darwin played an important role in shaping modern thought by proposing natural selection as the mechanism that drives evolution. Natural selection is an interaction between organisms and their environment that causes differential reproduction of different phenotypes and thereby alters the gene pool of a population. In essence the theory of evolution by natural selection is this:
1) In a population, there are heriTable differences between individuals.
2) HeriTable traits (alleles of genes) produce traits (phenotypes) that affect the ability of an organism to survive and have offspring.
3) Some individuals have phenotypes that allow them to survive longer, be healthier, and have more offspring than others.
4) Individuals with phenotypes that allow them to have more offspring will pass on their alleles more frequently than those with phenotypes that have fewer offspring.
5) Over time, those alleles that lead to more offspring are passed on more frequently and become more abundant, while other alleles become less abundant in the gene pool.
6) Changes in allele frequency are the basis of evolution in species and populations.
To put it simply, evolution occurs when natural selection acts on genetic variation to drive changes in the genetic composition of a population. A key term in evolution is fitness. In evolutionary terms, fitness is not how well an animal is physically adapted to a niche in the environment, or how well it can feed itself, but how successful it is in passing on its alleles to future generations. The way to have greater fitness is by having more offspring that pass on their alleles to future generations of the population. Some species achieve greater fitness through sheer numbers of progeny produced, who are then left to fend for themselves. Other species have fewer progeny, but protect and nurture the young to maturity.
• If an allele of a gene causes cancer in elderly polar bears after their reproductive years have passed, how will it affect the fitness of bears carrying the allele?92
• If a recessive allele causes sterility in homozygotes, how will it affect the fitness of heterozygotes?93
• A group of mice are infected with recombinant virus in bone marrow cells that allows the mice to live longer. The mice are then released into a wild population. Will natural selection act to increase the life span of the population?94
• Which of the following will have greater fitness: A fish that has two offspring and protects and nurtures its young to maturity, or a fish that has 10 offspring and abandons them, resulting in the death of 8 young fish before maturity?95
• The recessive allele that causes cystic fibrosis is strongly selected against in modern society, since individuals with this disease often die before sexual maturity. However, the frequency of the allele takes many generations to decrease in the population. Why?96
• A certain genetic disease is caused by a recessive allele. In the absence of effective therapy, homozygous individuals with this allele generally die before reaching sexual maturity. The allele also protects heterozygous individuals against several life-threatening viral diseases. If a medicine is found that provides a complete remedy for the disease, allowing individuals with the disease to live an entirely normal life, which of the following statements describes what will happen to the frequency of the allele in the population after that time?97
A) The frequency of the allele will decrease.
B) The frequency of the allele will remain constant.
C) The frequency of the allele will increase.
D) It is not possible to predict the future frequency of the allele.
Sources of Genetic Diversity
Natural selection acts on the genetic diversity in a population to alter allele frequencies, causing evolution. Genetic diversity in a population is a requirement for natural selection to occur. [If a population of sea otters contains only one allele of a gene that protects against cold, can natural selection drive evolution of this trait?98 Can natural selection cause new alleles to appear in the population?99] Natural selection does not introduce genetic diversity, however; it can act only on existing diversity to alter allele frequencies.
There are two sources of genetic variation in a population: new alleles and new combinations of existing alleles. New alleles are the result of mutations in the genome. New combinations of alleles are generated during sexual reproduction as a result of independent assortment, recombination and segregation during meiosis. By increasing and maintaining genetic variation in a population, sexual reproduction allows for greater capacity for adaptation of a population to changing environmental conditions.
• Do new alleles in a population generally confer greater or lesser fitness on an individual carrying them?100
• If a mutation occurs in a muscle cell of an individual who then has many progeny, does this mutation increase genetic variation in the population?101
• Does mitosis contribute to the genetic variation in a population?102
• If a population of flowers loses the ability to reproduce sexually and reproduces only asexually, how will this affect natural selection in the population?103
• Plants that are pollinated by insects sometimes have physical features of the flower that prevent self-pollination. What is the advantage to the plant of preventing self-pollination?104
• Which one of the following can create new alleles in a population?105
A) Non-random mating
B) Random drift
C) Recombination
D) Deletion
Modes of Natural Selection
Natural selection can occur in many different manners and have different effects in a population. The following are a few examples:
1) Directional Selection: Polygenic traits often follow a bell-shaped curve of expression, with most individuals clustered around the average and some members of a population trailing off in either direction away from the average. If natural selection removes those at one extreme, the population average over time will move in the other direction. Example: Giraffes get taller as all short giraffes die for lack of food.
2) Divergent Selection: Rather than removing the extreme members in the distribution of a trait in a population, natural selection removes the members near the average, leaving those at either end. Over time divergent selection will split the population in two and perhaps lead to a new species. Example: Small deer are selected for because they can hide, and large deer are selected because they can fight, but mid-sized deer are too big to hide and too small to fight.
3) Stabilizing Selection: Both extremes of a trait are selected against, driving the population closer to the average. Example: Birds that are too large or too small are eliminated from a population because they cannot mate.
4) Artificial Selection: Humans intervene in the mating of many animals and plants, using artificial selection to achieve desired traits through controlled mating. Example: The pets and crop plants we have are the result of many generations of artificial selection.
5) Sexual Selection: Animals often do not choose mates randomly, but have evolved elaborate rituals and physical displays that play a key role in attracting and choosing a mate. Example: Some birds have bright plumage to attract a mate, even at the cost of increased predation.
6) Kin Selection: Natural selection does not always act on individuals. Animals that live socially often share alleles with other individuals and will sacrifice themselves for the sake of the alleles they share with another individual. Example: A female lion sacrifices herself to save her sister’s children.
8.9 THE SPECIES CONCEPT AND SPECIATION
A species is a group of organisms which are capable of reproducing with each other sexually. (Other criteria, such as morphology, are used to classify species that only reproduce asexually.) [What’s the difference between a population and a species?106] Two individuals are not members of the same biological species if they cannot mate and produce fit offspring. [When a horse mates with a donkey a mule is born. Mules are healthy animals with long life spans, but they are sterile. Are horses and donkeys members of the same species?107] Reproductive isolation keeps existing species separate. There are two types of reproductive isolation: prezygotic and postzygotic.
Prezygotic barriers prevent the formation of a hybrid zygote. Such barriers may be:
• Ecological: individuals who could otherwise mate live in different habitats, and thus cannot access each other
• Temporal: individuals mate at different times of the day, season, or year
• Behavioral: some species require special rituals or courtship behaviors before mating can occur
• Mechanical: reproductive structures or genital organs of two individuals are not compatible (even if they court and attempt copulation)
• Gametic: sperm from one species cannot fertilize the egg of a different species due incompatibilities in the sperm-egg recognition system, discussed in Chapter 14
Postzygotic barriers to hybridization prevent the development, survival, or reproduction of hybrid individuals (those that arise from a mating between two different species), and thus prevent gene flow if fertilization between two different species does occur. There are three types of postzygotic barriers:
• Hybrid inviability: hybrid offspring do not develop or mature normally, and normally die in the embryonic stage
• Hybrid sterility: a hybrid individual is born and develops normally, but does not produce normal gametes, and thus is incapable of breeding (e.g., a mule, offspring of a mating between a horse and a donkey, is sterile)
• Hybrid breakdown: when two hybrids mate successfully to produce a hybrid offspring, but this second generation hybrid is somehow biologically defective
The creation of new species is known as speciation. An important premise in modern biology is that all species come from pre-existing species. Cladogenesis is branching speciation (clado is from the Greek for branch), where one species diversifies and becomes two or more new species. Anagenesis is when one biological species simply becomes another by changing so much that if an individual were to go back in time, it would be unable to reproduce sexually with its ancestors. One type of cladogenesis, allopatric isolation, is initiated by geographical isolation. Over time, geographical isolation leads to reproductive isolation. Sympatric speciation occurs when a species gives rise to a new species in the same geographical area, such as through divergent selection.
Cladogenesis has left traces which taxonomists use to classify organisms. Homologous structures are physical features shared by two different species as a result of a common ancestor. For example, bird wings have five bony supports which resemble distorted human fingers, and dog paws also resemble distorted human hands. The explanation is that dogs, birds, and people all have a common ancestor which had five-toed feet. Analogous structuresserve the same function in two different species, but not due to common ancestry. The flagellum of the human sperm and bacterial flagella are an example; they have entirely different structures from different organisms yet play the same role in motility. Convergent evolution is when two different species come to possess many analogous structures due to similar selective pressures. For example, bats and birds appear very similar even though bats are mammals. The opposite of convergent evolution is divergent evolution, in which divergent selection causes cladogenesis. Parallel evolution describes the situation in which two species go through similar evolutionary changes due to similar selective pressures. For example, in an ice age, all organisms would be selected for their ability to tolerate cold.
8.10 TAXONOMY
Taxonomy is the science of biological classification, originated by Carolus Linnaeus in the eighteenth century. He devised the binomial classification system we use today, in which each organism is given two names: genus and species. The binomial name of an organism is written in italics (or is underlined) with the genus capitalized and the species not, as in Homo sapiens (man the wise). There are eight principal taxonomic categories: domain, kingdom, phylum, class, order, family, genus, and species.108 You should know how humans are classified and the defining characteristics of each category. Table 2 below provides a general summary. Table 3 on the next page summarizes human taxonomy.
Table 2 Taxonomic Characteristics
Domain Bacteria and Domain Archaea were both previously classified into Kingdom Monera. Because of huge diversity in these organisms, they have since been separated into two domains, and for these organisms, the kingdom and domain are the same.
Table 3 Human Taxonomy
Regarding subphylum Vertebrata: Bilateral symmetry is the same as plane symmetry, where the organism has an internal line of symmetry and two halves that are mirror images. Humans, cats, and butterflies are all examples. Bilateral symmetry is associated with cephalization (an evolutionary trend, where nervous system tissue becomes concentrated at one end of the organism, and can eventually become a head with sensory organs) and more efficient locomotion. Not all animals have bilateral symmetry. Some primitive animals (such as sponges) are asymmetrical, and others (such as starfish and jellyfish) have radial symmetry, where the animal resembles a pie, and each cross section of a pie piece looks the same.
Many animals have some sort of skeleton to support their body and help with movement. An exoskeleton is on the exterior, whereas an endoskeleton is found inside. Arthropods (such as insects, spiders, and crustaceans like lobsters) have a hard exoskeleton made of chitin109. Some chordates have a flexible endoskeleton made of cartilage, while others have stronger endoskeletons made of bone. Table 4 and Figure 19 contain additional details on subphylum Vertebrata.
Table 4 Characteristics of the Vertebrate Classes
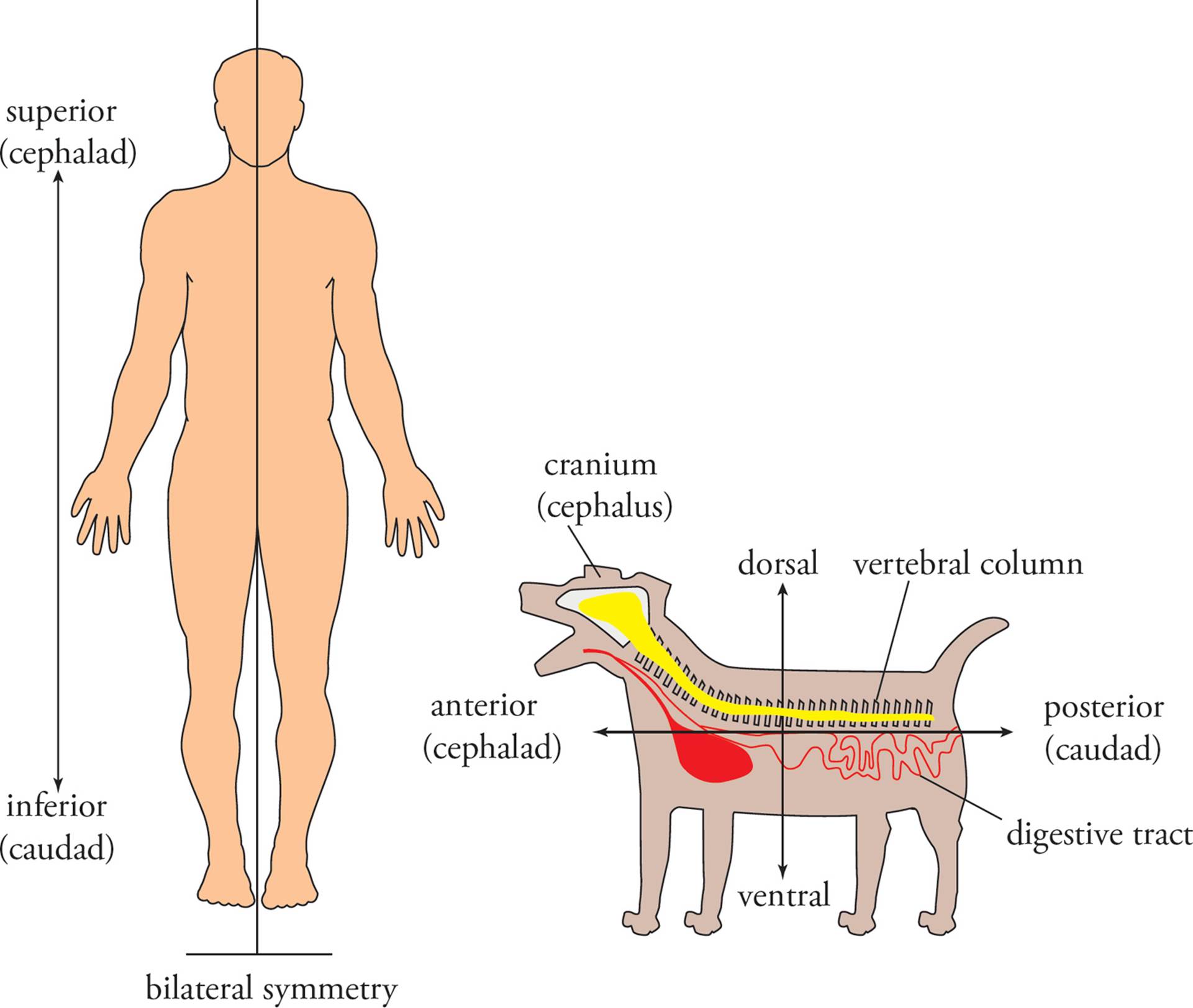
Figure 19 Bilateral Symmetry and the Anatomical Axes
Anatomists describe bodies with reference to axes such as the dorsal-ventral axis and the anterior-posterior axis (Figure 19). These are imaginary planes through the body. Anterior means “front-facing”; posterior is the opposite. Dorsal means “on top.” A shark has a “dorsal fin,” and the spines of dogs and humans are considered dorsal as well. Ventral is the opposite of dorsal. The bellybutton is a ventral structure in dogs and humans. In humans, superioris used to indicate “toward the head.” Inferior means “toward the feet.” Another way to say “toward the head” is cephalad. (Cephalus is Latin for head.) The opposite is caudad, meaning “toward the tail.” [Does the axis of bilateral symmetry in vertebrates run parallel or perpendicular to the dorsal-ventral axis?110]
8.11 THE ORIGIN OF LIFE
Based on radioisotope dating, the earth is thought to be 4.5 billion years old. All life evolved from prokaryotes. The oldest fossils are 3.5 billion-year-old outlines of primitive prokaryotic cell walls found in stromatolites (layered mats formed by colonies of prokaryotes). Even older life forms certainly existed, but lacked cell walls and thus left no fossil record (at least none have yet been discovered). Hence life on Earth is older than 3.5 billion years, nearly as old as the planet itself.
The atmosphere of the young Earth was different from today’s atmosphere. The predominant gases then were probably H2O, CO, CO2, and N2. The most important thing to note here is the absence of O2. It is thought that the early atmosphere was a reducing environment, where electron donors were prevalent (see Chapter 3). Oxygen is an electron acceptor, and as such tends to break organic bonds. In this early world, simple organic molecules, or monomers (“single units”) could form spontaneously. The energy for this synthesis was provided by lightning, radioactive decay, volcanic activity, or the Sun’s radiation, which was more intense than it is today due to the thinner atmosphere. Laboratory recreations of the early environment result in the spontaneous formation of amino acids, carbohydrates, lipids, and ribonucleotides, as well as other organic compounds.
Spontaneous polymerization of these monomers can also be observed in the lab (including spontaneous polymerization of ribonucleotides). No enzymes were present when this was occurring for the first time in nature, but it is thought that metal ions on the surface of rocks and especially clay acted as catalysts. This is known as abiotic synthesis. Polypeptides made in this way are called proteinoids.
Proteinoids in water spontaneously form droplets called microspheres. When lipids are added to the solution liposomes form, with lipids forming a layer on the surface of proteins. A more complex particle known as a coacervateincludes polypeptides, nucleic acids, and polysaccharides. Coacervates made with pre-existing enzymes are capable of catalyzing reactions. Microspheres, liposomes, and coacervates are collectively referred to as protobionts.
Protobionts resemble cells in that they contain a protected inner environment and perform chemical reactions. They can also reproduce to a certain extent: when they grow too large they split in half. What is lacking, however, is an organized mechanism of heredity. This was first provided by RNA. As noted above, RNA chains form spontaneously in the appropriate solution. Even more interesting is the observation that single-stranded RNA chains can be self-replicating. A daughter chain lines up on the parent by base pairing and then spontaneously polymerizes with a surprisingly low error rate. A nonspecific catalyst such as a metal ion can further increase the efficiency of RNA self-replication. Furthermore, it is now known that RNA has catalytic activity in modern cells. For example, in primitive eukaryotes, introns are spliced out of the mRNA by ribozymes, which are RNA enzymes.
Somehow a mechanism evolved for polypeptides to be copied from early RNA genes. You already know about the inherent tendency for phospholipids to form lipid bilayers. Given all this information, it’s not too hard to imagine true cells evolving from a primordial soup at the dawn of time. The last step in the evolution of the earliest cells would have been the switch from RNA to DNA as the genetic material. DNA is more stable due to its 2′-deoxy structure and also due to the fact that it spontaneously forms a compact double-stranded helix.
Chapter 8 Summary
• Organisms express phenotypes (physical characteristics) according to their genotypes (combinations of alleles).
• From a single diploid precursor cell, meiosis generates four haploid cells (gametes) with a random mix of alleles. This is due to crossing over in prophase I, and separation of homologous chromosomes in anaphase I. Nondisjunction is a failure to separate the DNA properly during meiosis, and can result in gametes with improper numbers of chromosomes.
• The Punnett square or the rules of probability can be used to determine the genotypes and phenotypes of offspring from given crosses, or the probability of having offspring with certain traits.
• The rule of multiplication states that the probability of A and B occurring is equal to the probability of A multiplied by the probability of B.
• The rule of addition states that the probability of A or B occurring is equal to the probability of A plus the probability of B, minus the probability of A and B together.
• Classical dominance occurs when a phenotype or trait is determined by one gene with two alleles, and one allele is dominant (expressed) and the other is recessive (silent). There are several exceptions to classical dominance, including incomplete dominance, codominance, epistasis, pleiotropism, polygenism, and penetrance.
• Incomplete dominance occurs when two different alleles for a single trait result in a blended phenotype. Codominance occurs when two different alleles for a single trait are expressed simultaneously, but independently (no blending).
• Epistasis occurs when the expression of one gene depends on the expression of another.
• Pleitropic genes affect many different aspects of the overall phenotype, while polygenic traits are affected by many different genes.
• Penetrance refers to the likelihood that a particular genotype will result in a given phenotype. Penetrance can be affected by several factors including age, environment, and lifestyle.
• Linkage occurs when two genes are close together on the same chromosome; it leads to alleles being inherited together (less recombination) instead of independently.
• Pedigrees can be used to analyze the patterns of inheritance of different traits. There are six primary modes of inheritance: autosomal recessive, autosomal dominant, mitochondrial, Y-linked, X-linked recessive, and X-linked dominant.
• The Hardy-Weinberg law can be used to study population genetics. It assumes classical dominance with only two alleles and unchanging allele frequencies. It is based on five assumptions: no mutation, no natural selection, no migration, large populations, and totally random mating.
• Natural selection drives evolution by allowing individuals with random, beneficial mutations to survive and pass those beneficial mutations on to their offspring.
• Homologous structures are the result of divergent evolution to form new species, and analogous structures are the result of convergent evolution, in which different start species must meet similar environmental challenges.
CHAPTER 8 FREESTANDING PRACTICE QUESTIONS
1. A woman is phenotypically normal but had a brother who had an autosomal recessive disorder that resulted in death during infancy. What is the probability that this woman is a carrier for the disorder that afflicted her brother?
A) 1/4
B) 1/3
C) 1/2
D) 2/3
2. A set of inherited traits show a phenotypic ratio of 9:3:3:1 among offspring of a given mating. A possible explanation for this observation is a two locus–two allele system where each locus exhibits:
A) independent assortment.
B) linkage.
C) epistasis.
D) incomplete dominance.
3. If a woman is a carrier for an X-linked recessive disorder and mates with a normal, unaffected male, what is the probability her grandson has the disorder? Assume she has a normal son and a daughter, and they both have homozygous normal partners.
A) 0
B) 1/4
C) 1/2
D) 3/4
4. A geneticist is analyzing a family tree, where mothers affected by the trait under study pass the trait on to 100% of their offspring, both sons and daughters. Fathers affected by the trait pass it on to 0% of their offspring. All affected individuals have an affected mother. What is the inheritance pattern of this trait?
A) Y-linked
B) Autosomal dominant
C) Autosomal recessive
D) Mitochondrial
5. A purebred long-tailed cat with long whiskers (TTww) is mated to a purebred short-tailed cat with short whiskers (ttWW). The kittens will be:
A) 100% long-tailed with long whiskers.
B) 100% long-tailed with short whiskers.
C) 100% short-tailed with short whiskers.
D) 50% long-tailed cat with long whiskers, 50% short-tailed cat with short whiskers.
6. A homozygous white bull is mated to a homozygous cow with a red coat. The offspring all have a roan coat color, which is composed of a mix of white hairs and red hairs. Which of the following is true?
A) Coat color in cattle is an example of codominance.
B) White hair is recessive to red hair.
C) Coat color in cattle is an example of incomplete dominance.
D) White hair is dominant over red hair.
7. If the genes for ear size (E or e) and aggressiveness (A or a) in mice are linked and 25 mu apart, what is the probability of getting a large-eared aggressive pup (EeAa) from a mating between a small-eared aggressive female (eeAA) and a large-eared nonaggressive male (EEaa)?
A) 12.5%
B) 25%
C) 75%
D) 100%
CHAPTER 8 PRACTICE PASSAGE
The fruit fly, Drosophila melanogaster, is an ideal organism on which to study genetic mechanisms. This organism has simple food requirements, occupies little space, and the reproductive live cycle is complete in about 12 days at room temperature, allowing for quick analysis of test crosses. In addition, fruit flies produce large numbers of offspring, which allows for sufficient data to be collected quickly. Many Drosophila genes are homologous to human genes, and are studied to gain a better understanding of what role these proteins have in humans.
To understand the inheritance patterns of certain genes in Drosophila, the following experiments were carried out. Assume that the alleles for red eyes, brown body, and normal wings are dominant and the alleles for white eyes, ebony body, and vestigial wings are recessive.
Experiment 1:
A red-eyed female and a white-eye male were crossed. The subsequent generation of flies was also crossed. The phenotypic results of both generations are shown below:
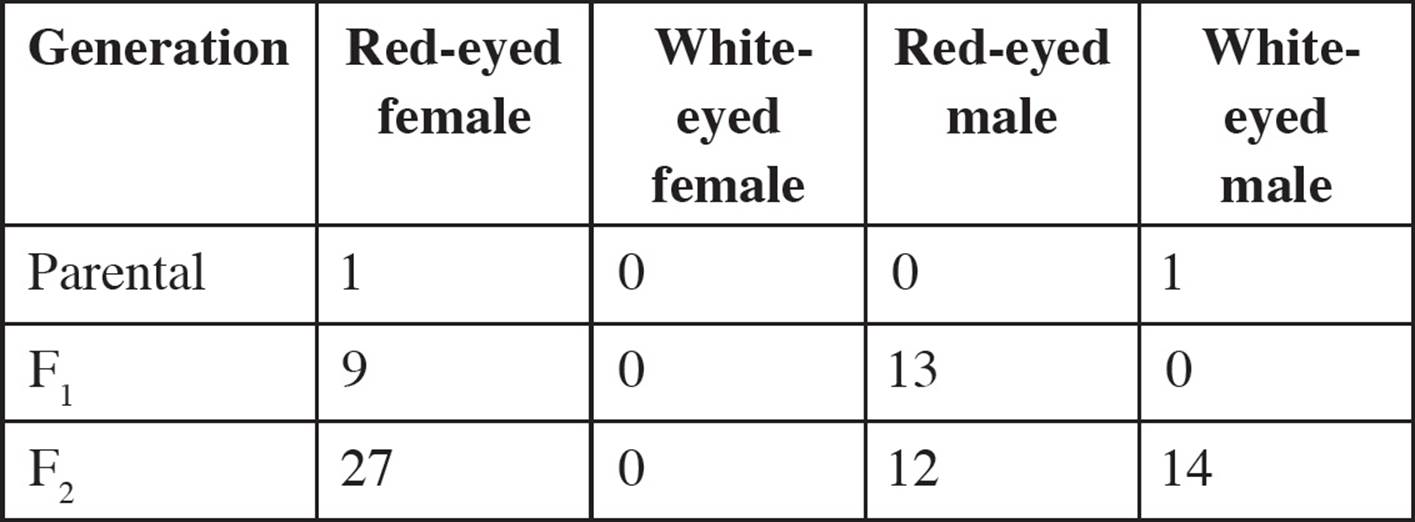
Experiment 2:
Five of the red-eyed female Drosophila from the F1 generation of Experiment 1 were crossed with white-eyed males. The result of this cross is shown below:
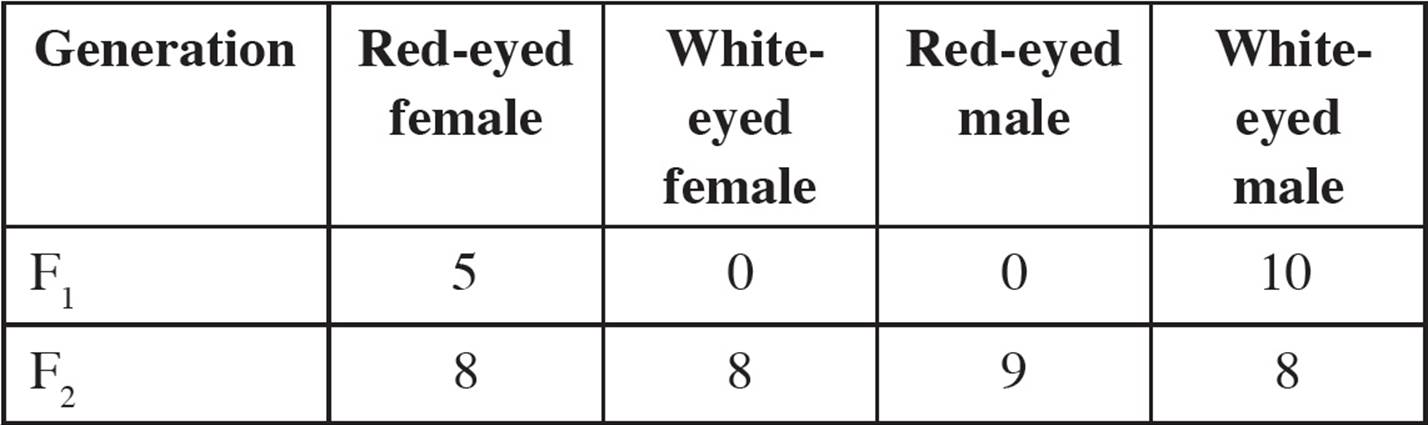
Experiment 3:
The white-eyed females from Experiment 2 were crossed with red-eyed males. The result of this cross is shown below:
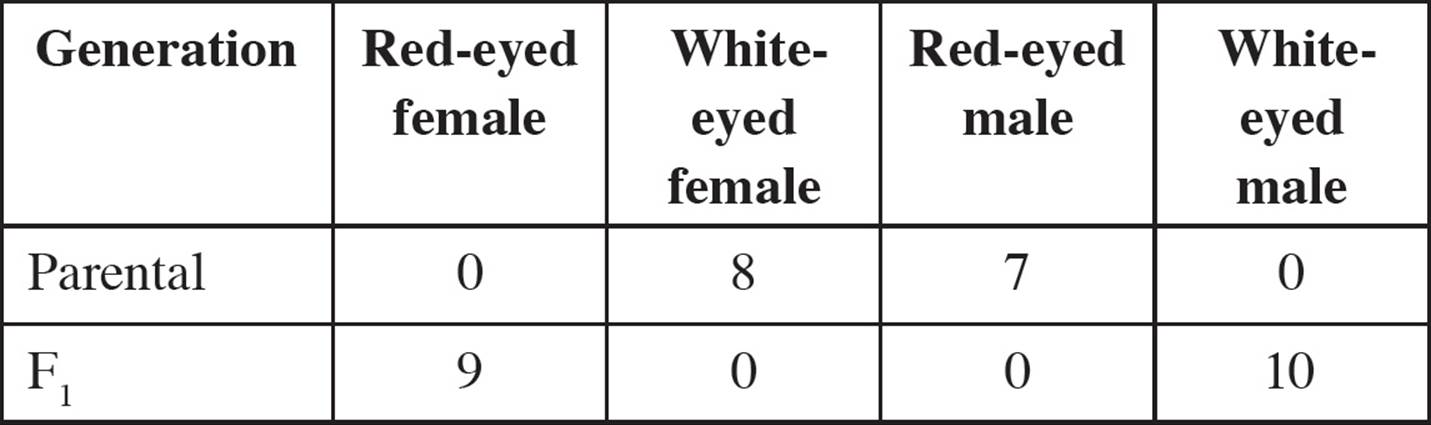
Experiment 4:
A male heterozygous for body color and wing type is crossed with an ebony, vestigial-winged female. The results of the cross are shown below:
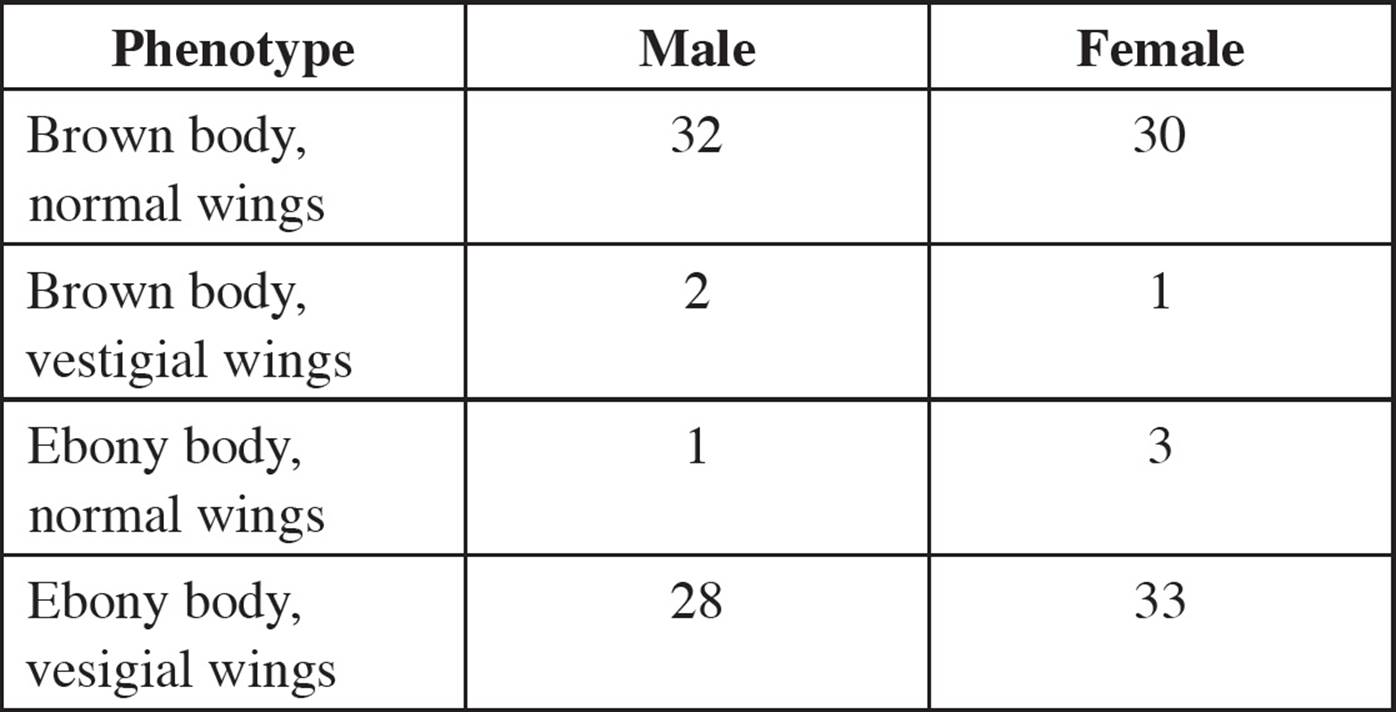
1. What is the most likely mode of inheritance for white eye color?
A) X-linked recessive
B) X-linked dominant
C) Autosomal recessive
D) Autosomal dominant
2. What ratio of white-eyed to red-eyed females would be expected if the F1 generation of Experiment 3 were crossed?
A) White-eyed females would not be present.
B) Red-eyed females would not be present.
C) 1:1
D) 1:2
3. Approximately how many of the red-eyed females of the F2 generation of Experiment 2 are homozygous dominant?
A) 0
B) 2
C) 4
D) 8
4. Which of the following statements is/are true with regard to the results obtained from Experiment 4?
I. The genes for body color and wing type are on the same chromosome.
II. Recombination occurred.
III. The heterozygous male had a dominant and recessive allele on each homologous chromosome.
A) I only
B) I and II
C) I and III
D) II only
5. What is the recombination frequency shown by the results of Experiment 4?
A) Recombination did not occur.
B) 5%
C) 50%
D) 95%
6. Assume that short legs are due to a recessive allele on a chromosome other than body color and that dominant alleles are considered wild-type. A white-eyed male is mated to an ebony colored, short-legged female and the F1 males resulting are wild type. If these males are backcrossed to ebony-colored, short-legged females, what proportion of the F2 offspring will be wild-type males?
A) 0
B) 1/2
C) 1/4
D) 1/8
7. If the frequency of vestigial wings in a population of Drosophila (n = 1500) is 4%, how many flies would be heterozygous with regard to wing type? Assume random mating, but no migration, mutation, random drift, or natural selection.
A) 60
B) 240
C) 480
D) 960
SOLUTIONS TO CHAPTER 8 FREESTANDING PRACTICE QUESTIONS
1. D If the woman’s brother was affected with this autosomal recessive disorder, both of her parents must have been carriers for the trait. Since it is lethal in infancy, you know both parents must have been heterozygous. The mating between two heterozygotes produces the following genotypic ratio in their offspring: 25% homozygous recessive, 50% heterozygous, 25% homozygous dominant. Since the woman is phenotypically normal, she must not have the disorder, so she is not in the 25% homozygous recessive group. She is either in the 25% homozygous dominant group or the 50% heterozygous group, so the chance she is a carrier is 2/3 (choice D is correct).
2. A If two heterozygous parents are mated (for example, AaBb × AaBb), the expected F1 phenotype ratio if the two traits are assorting independently is 9:3:3:1 (choice A is correct). If linkage were occurring, this ratio would be skewed after the mating of two double heterozygotes, since alleles would be preferentially inherited together. Since the ratio is not skewed, linkage is not occurring (choice B can be eliminated). This is not a case where one gene is silencing another, or controlling the expression of the other, as this would also lead to a skewed ratio (choice C can be eliminated). Incomplete dominance of two alleles leads to a blended heterozygous phenotype; thus, if it occurs at a single locus, it produces three different phenotypes. If it were to occur at two different loci (as in this case), there would be nine possible phenotypes (3 possible phenotypes at the first locus times 3 possible phenotypes at the second locus = 32 = 9). In this case there are only four different phenotypes (choice D can be eliminated).
3. B If we assign “A” as the normal allele and “a” as the affected allele, the woman is XAXa and her mate is XA Y. The question stem says that she has a normal son, who must have the genotype XA Y; he received the XA from his mother and his Y chromosome from his father. There is no probability associated with this that must be factored into the solution, because this is the only way the couple could have a normal son. If this son mates with a homozygous normal female, he will pass his Y chromosome onto his son(s), which means there is zero chance of having an affected son. In other words, an XA Y × XA XA mating cannot generate Xa Y sons. Next, let’s work with the daughter of the woman in the question stem. The question says she has a normal phenotype. She must receive an XA from her father. There is a 50% probability she will receive an Xa from her mother. If the daughter is XAXa (remember there is a 50% chance of this) and her mate is XA Y, there is a 50% chance their son will be affected. Overall, then, the probability that a grandson is affected is 0 + (1/2)(1/2), or 1/4. Therefore, choice B is correct.
4. D Females are not affected by Y-linked traits (choice A is wrong). If the trait was autosomal dominant, affected fathers would pass the trait to at least some of their offspring (choice B is wrong). Autosomal recessive traits do not demonstrate the sex bias that is described in the question stem (choice C is incorrect). By process of elimination, this trait must be inherited via the mitochondrial genome. All humans inherit their mitochondrial genome from their mothers; during fertilization, the sperm only contributes 23 chromosomes but the ovum contributes 23 chromosomes along with all other cellular organelles and cytoplasm (choice D is correct).
5. B The offspring will be 100% TtWw and will display the dominant phenotype for each trait. Since each parent has a dominant and a recessive phenotype, the kittens will not have the phenotype of either parent (choice B is correct and choices A, C, and D are wrong).
6. A Since the offspring are neither red nor white, this trait is not inherited via simple Mendelian genetics, and neither trait is dominant or recessive (choices B and D are wrong). The individual hairs on the F1 cattle are either red or white, which means that both alleles are being expressed. This best matches the definition of codominance (choice A is correct). If the individual hairs were a blend of white and red, the best answer would have been incomplete dominance (choice C is wrong).
7. D The pups will get eA from their mom and Ea from their dad. All the pups will be eA/Ea, so the answer must be D. Note that the linkage information was not useful in answering this question.
SOLUTIONS TO CHAPTER 8 PRACTICE PASSAGE
1. A The passage states that the allele for red eyes is dominant and the allele for white eyes is recessive (choices B and D are wrong). The results from Experiments 1 suggest that the gene for eye color is X-linked. If it were autosomal, all F1 flies from that experiment would be heterozygous, and there would be an equal distribution of both genders and colors in the F2 generation. Remember that a gender bias in the phenotype of a trait usually indicates that you’re working with a sex-linked trait.
2. C The passage states that the red allele is dominant and the white allele is recessive; further eye color is a sex-linked trait (see the explanation for Question 1). For Experiment 3, the female parental genotype must be homozygous recessive (XcXc), and the male parental genotype must be XC Y. Thus, the F1 generation consists of heterozygous females (XC Xc) and males with the recessive white-eyed allele (Xc Y). If these two were mated with each other, the female progeny of this generation would consist of homozygous recessive (XcXc) and heterozygous (XC Xc) females in an approximate 1:1 ratio (choice C is correct).
3. A Experiment 2 is a cross between an F1 female and a white-eyed male. The genotype of the F1 females from Experiment 1 is XC Xc; they have red eyes, so they must have XC, and they are female, so they inherited Xc from their white-eyed father. If these XCXc females are crossed with Xc Y (white-eyed) males, the females generated in the F2 must be 50% XC Xc (red-eyed) and 50% XcXc (white-eyed). If the father has white eyes, it is impossible to generate homozygous red-eyed females in the F2 (choice A is correct).
4. B There is no sex bias in the results from Experiment 4, so we can assume the alleles are autosomal. The passage states that the alleles for brown bodies and normal wings are dominant, and the alleles for ebony bodies and vestigial wings are recessive. Let’s assign B = brown and b = ebony for body color, and W = normal and w = vestigial for wing phenotype. Thus the cross in Experiment 4 is BbWw × bbww. Item I is true: If the genes for body color and wing type were on separate chromosomes, one would expect the law of independent assortment to hold true and the phenotypic ratio of the progeny would be expected to be 1:1:1:1. However, this is not the case. Two of the phenotypes predominate, indicating that the genes are close together on the same chromosome, or are linked (choice D can be eliminated). Item II is also true: The recombinant phenotypes, although rare, are nonetheless seen. Recombination between homologous chromosomes is the best explanation for this observation (choices A and C can be eliminated and choice B is correct). Note that Item III is false: The double heterozygous Drosophila must have both dominant alleles on one chromosome and both recessive alleles on the other to account for the results seen. In other words, the cross performed was BW/bw × bw/bw. The parental combinations of alleles are BW and bw (more frequent), and the recombinant combinations of alleles (less frequent) are Bw and bW.
5. B Recombination did in fact occur (see explanation for Question 4; choice A is wrong). Recombination frequency = number of recombinants/total number of offspring. In this case, the total number of recombinants is 7 and the total number of offspring is 130. 7/130 = 0.05 or 5% recombination frequency (choice B is correct and choices C and D are wrong). Note that choice D is the frequency of non-recombination, and choice C would be correct if the genes were not linked.
6. D Similar to above, let’s assign XC = red eyes (dominant and wild type), Xc = white eyes, B = brown body color (dominant and wild type), b = ebony body color, and now S = long legs (dominant and wild-type), and s = short legs. The first cross in the question stem is: XcYBBSS × XC X Cbbss. Note that if the phenotype is not listed, we can assume it is wild-type. Also, since all the male offspring from this cross are wild-type, then the male parent must have been homozygous for body color and leg length, and the female parent must have been homozygous for eye color. The male F1 offspring will be X CYBbSs. Next, this male was backcrossed to a female from the previous generation (X CYBbSs × X CX Cbbss), and we are asked for the probability of a wild-type male. This is equal to the proportion that they are male (1/2), multiplied by the proportion that they will have brown body color (1/2), multiplied by the proportion that they will have long legs (1/2). Therefore, the proportion that are wild type = 1/2 × 1/2 × 1/2 = 1/8 (choice D is correct). Notice that we do not have to factor in the probability of the Drosophila being red-eyed because this is guaranteed given the genotype of their mother.
7. C Using the Hardy-Weinberg equations for allele frequency and genotype, the frequency of the vestigial wing allele (q) must be equal to √0.04, or 0.20. Therefore, the frequency of the normal wing allele (p) is 0.80 and the frequency of the heterozygous genotype (pq and qp) is equal to 2 × 0.2 × 0.8, or 0.32. The actual number of Drosophila that would have this genotype would be 480 (0.32 × 1500).
1 No. tRNA and rRNA genes, as well as other small nuclear RNA genes, do not encode polypeptides.
2 No. Every gene is located at a specific locus, but physical traits, particularly complex traits, like weight or height, can be controlled by many different genes and therefore do not map to a single locus, but to many.
3 Yes, there can be many versions (alleles) of a particular gene. Under normal circumstances, however, one individual cannot have more than two of those different alleles, since they have only two copies of a gene (one on each homologous chromosome). An exception is when an individual is polyploid for a certain chromosome (i.e., they have more than two homologous chromosomes, for example in Down syndrome and Klinefelter syndrome).
4 An individual with two different alleles at a given locus has one allele on one chromosome and the other allele on its homologous partner (so D is correct and choice B is not possible). While choice A may be possible, it is an exceedingly complex phenomenon and not discernible from the information given. The question discusses a single individual, not a pair of siblings (eliminating choice C).
5 No. If there is only one copy of a gene, then that is the copy which determines the phenotype.
6 CC and Cc individuals have curly hair, and cc individuals have straight hair. Only homozygous recessive individuals express recessive traits. In the heterozygote, the presence of the recessive allele is masked by the dominant allele, so there are only two different phenotypes, although there are three different genotypes. This type of interaction between alleles is called classical dominance.
7 The organism is diploid normally, with two copies of two chromosomes, or four chromosomes total. After DNA synthesis, during prophase I, the cell still has four chromosomes; however, the chromosomes are replicated and held together at the centromere. Thus each chromosome consists of two sister chromatids, and the cell has a total of eight sister chromatids.
8 Not if it is done correctly. Error free recombination involves a one-for-one swap of DNA between homologous chromosomes.
9 Yes. Although each chromosome contains the same genes after crossing over, it may contain different alleles of some genes that were not present on the same chromosome previously.
10 No. Homologous chromosome separate (segregate) randomly. This is one aspect of meiosis that increases genetic variation during sexual reproduction.
11 The sister chromatids would be identical, except that recombination with homologous chromosomes occurred earlier in meiosis, during prophase I, altering the sister chromatids.
12 Item I is false: The spindle separates sister chromatids during both (choices A and C can be eliminated.) Note that both remaining answer choices include Item II, so Item II must be true. Item II is true: Only meiosis involves pairing and recombination between homologous chromosomes. Item III is false: Meiotic recombination occurs between homologous chromosomes, not sister chromatids (choice D can be eliminated and choice B is correct).
13 Crossing over occurs during prophase I, separation of homologous chromosomes occurs during anaphase I, and nuclear envelope breakdown occurs during prophase I and sometimes prophase II (choices A, B, and D are false). Only separation of sister chromatids occurs after metaphase II, in anaphase II. Answer: C.
14 Normal gametes have one copy of each chromosome; this is the definition of haploid.
15 If the homologous chromosomes do not separate in meiosis I, then one daughter cell from this division will have four copies of this chromosome and the other cell will have none. In meiosis II, sister chromatids will separate, leaving two gametes with two copies of the chromosome and two gametes with no copies of the chromosome.
16 There is no information missing in a person with trisomy. All of the chromosomes are present, and there is no reason to believe that any of the genes on these chromosomes are deleted or mutated to render them inactive.
17 The problem with trisomy appears not to be that genetic information is missing, but that there is too much present. A mechanism involved might be gene dosage. Genes are regulated to produce the right amount of each gene product. In trisomy, many genes are present in one more copy than usual, resulting in greater quantities of the gene products encoded on this chromosome. The extra quantities of so many gene products, even if they are normal in sequence, can have dramatic consequences.
18 During meiosis I, at the time when homologous chromosome separate.
19 If a strain always produces the same trait when mated with itself, it is likely to be homozygous for the trait. The pure-breeding yellow pea is homozygous for the yellow allele g of the color gene.
20 The two strains were both pure-breeding and could only produce gametes containing one type of allele. All of the progeny would be the Gg genotype. If all progeny are green, then the green allele is dominant and the yellow allele is recessive.
21 No. A green plant could either be heterozygous Gg or homozygous GG.
22 The original pea is heterozygous Gg.
23 The gamete must be the result of nondisjunction.
24 Yes. The principles are the same for human genes as for pea genes, as long as an organism is diploid and goes through sexual reproduction.
25 No, they would not. They would display an important exception to independent assortment, linkage, which is discussed later.
26 According to independent assortment, the segregation of one gene does not depend on segregation of another. The chances of a gamete containing the W allele are 50%, regardless of the identity of the color allele.
27 If G (green allele) is dominant, then both GG homozygotes and Gg heterozygotes will be green, while only gg homozygotes will be yellow. 25% of the offspring in Figure 11 will be GG homozygotes, and 50% will be Gg heterozygotes, so a total of 75% of the offspring will be green (choice C).
28 Plant 1 has green wrinkled peas, while Plant 2 has green smooth peas.
29 All peas receive one w allele and one W allele, so all are wrinkled Ww heterozygotes (i.e., 0% are smooth).
30 All F1 peas are wrinkled and 75% are green, so 75% are green and wrinkled.
31 All F1 peas are wrinkled and 25% are yellow, so 25% are yellow and wrinkled.
32 The results obtained in reality rarely agree exactly with the predicted result. If the results differ slightly from the prediction, the most likely explanation is statistical variability. The more progeny from the cross, the closer the result should be to the prediction.
33 Independent assortment and the principle of segregation are inherent in the Punnett square. There is no reason to believe that these are not followed, making choice B the best response. Choices A, C, and D all assume that either independent assortment or segregation did not occur.
34 There are two different genotypes possible for the green wrinkled phenotype in the F1 generation: GGWw or GgWw. The best way to determine all possible phenotypes in the cross is to draw a Punnett square for both of these potential genotypes:
If the F1 plant is GGWw: If the F1 plant is GgWw:
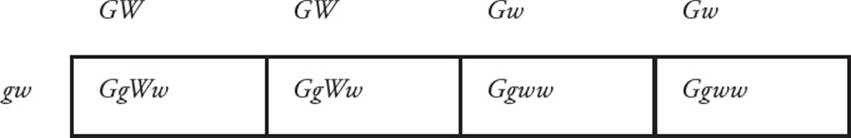
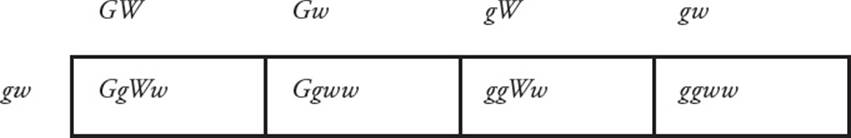
35 If yellow smooth progeny are observed, the F1 plant must be GgWw.
36 Without drawing a Punnett square, it is possible to see that all children must receive at least one b allele (from the father), and that 50% of the children will receive the B allele from the mother; thus, 50% of the children will be Bb. The odds of a boy are 50%. Therefore, the odds a child is both a boy and has the Bb genotype are, by the rule of multiplication, 0.5 × 0.5 = 0.25, or 25%.
37 Rr heterozygotes will have the phenotype of the dominant allele: red.
38 In this case, RW heterozygotes will be neither red nor white, but a blend of the two: pink.
39 Three phenotypes and three genotypes: RR (red), RW (pink), and WW (white).
40 The red blood cells will express both type A and type B antigens, so the blood type will be AB.
41 The red blood cells will express type B antigen only, and the blood type will be B.
42 Because they are both heterozygous, the woman’s genotype is IAi and the man’s genotype is IBi. Thus, their children could be IAIB (type AB), IAi (type A), IBi (type B), or ii (type O).
43 The trait of cancer development is probably polygenic, so it does not display simple patterns of inheritance. Cancer development is also influenced by the environment, such as exposure to carcinogens, further complicating the penetrance of the genotype.
44 The key variable must not lie within the allele itself, since this remains the same (so A and D are wrong). The genetic background of the two different strains of mice must affect whether or not the heart defect phenotype is expressed. Further, only one defect is observed (so it can’t be pleitropic; choice C is wrong). Therefore, the heart defect phenotype must be influenced by some other locus that is different in the two strains of mice, making B the best choice.
45 In males, recessive alleles on the X chromosome are always expressed, since no other allele is present that can mask the recessive allele.
46 Yes, they will. Assortment of nonhomologous chromosomes into gametes is random during meiosis.
47 A translocation occurs when a piece of one chromosome is moved onto another chromosome. The two genes are then found on the same chromosome and may not assort independently.
48 Yes. A TTgg individual can only make Tg gametes, regardless of whether the genes are on the same chromosome or not.
49 No. To predict how these traits will assort, it is necessary to know which alleles are present together on the same chromosome.
50 There are only two phenotypes: 50% tall green and 50% short yellow.
51 The pure-breeding short yellow plant can only have tg gametes. The tall green plant can make only two types of alleles, the same gametes shown for its parent. The results of the backcross will be the same as for the original cross in Figure 15, with 50% tall green and 50% short yellow plants.
52 The ttgg individual can make only tg gametes. The TtGg individual can make four different types of gametes if the genes are not linked: TG, Tg, tG, and tg. The genotypes and phenotypes of the cross will be 25% TtGg (tall green), 25% Ttgg (tall yellow), 25% ttGg (short green), and 25% ttgg(short yellow).
53 The gene for shape (wrinkled vs. smooth) is on a different chromosome than the other two genes, so there is no correlation between the wrinkled trait and the other traits (eliminating choices A and B). To be yellow, a pea must be homozygous gg. One of the g alleles must come from the chromosome with T and g together, making all yellow plants tall (choice C is correct). Some of the Tg gametes will join with tG gametes from the ttGg individual, meaning that some plants are tall and green (choice D is wrong). Drawing a Punnett Square can help to solve this problem.
54 ABC or abc genotypes
55 Only one phenotype. The F1 generation will all receive a TG chromosome from the pure-breeding tall green parent and a tg chromosome from the pure-breeding short yellow parent.
56 75% tall green and 25% short yellow. Try a Punnett square to verify this result, remembering to assort alleles together into gametes.
57 9:3:3:1 of tall green, tall yellow, short green, and short yellow, respectively. This is a classical Mendelian ratio observed when heterozygotes at two alleles are crossed.
58 The tall yellow and short green phenotypes would not be observed if linkage was complete (as in #56). The only way to produce these phenotypes is if a small number of gametes received chromosomes in which the TG and tg alleles were separated from each other by recombination, so these are the recombinant phenotypes.
59 If the genes are on the same chromosome, but are far apart from one another, then recombination occurs frequently. The genes will assort randomly during meiosis and will not display any linkage even though they are on the same chromosome.
60 The farther two genes are away from each other, the greater the odds that recombination will occur between them.
61 The F1 progeny are all BbGg genotype and therefore are big flower green plants.
62 The small flower green phenotype could not be produced by recombination. These alleles do not exist together in either parent and so could not be recombined together in the gametes. This must be the result of mutation.
63 Big flower yellow plants and small flower green plants can only be produced through recombination between the flower size and color genes.
64 The frequency of recombination is 16 recombinant phenotypes out of 100 progeny = 16%.
65 The maximal frequency of recombination would be when there as no linkage and the genes assorted independently. In this case there would be 25% big flower yellow plants and 25% small flower green plants, or 50% maximal frequency of recombination.
66 There is less recombination between the height and flower size genes (10% frequency) than between the color and flower size genes (16%), so the height gene is closer to the flower size gene than the color gene is.
67 The flower size and color genes have the most recombination between them, so they must be farthest apart, with the height gene in the middle.
68 Yes, but it requires one or more genes located between the two genes. The distance between the two genes could not be mapped directly by measuring the frequency of recombination between them, but if the distance from both of them to a gene in the middle can be mapped, then the overall distance between the genes can be mapped. Whole chromosomes can be mapped this way.
69 Item I is true: The simplest explanation is that the grandparent on each side passed on the fragile allele and the blond allele, but recombination occurred, so that the fragile and blond alleles assorted independently. Item II is false: Jose and Tonya both have the dominant sturdy bone gene in at least one copy, so some children are likely to have sturdy bones. Item III is true: Both Jose and Tonya may have one chromosome with the blond allele and the fragile bone allele linked. If so, a nonrecombinant phenotype would be blond/fragile. The answer is C.
70 No. Females never have a Y chromosome and so can never carry or express a Y-linked trait.
71 No. Y-linked traits are carried in only one copy, since there is only one Y chromosome per cell. If a male carries a recessive Y-linked trait, he will express it.
72 Since males receive their X chromosome from their mother (and their Y chromosome from their father), they receive X-linked traits from their mother.
73 An allele that encodes inactive protein or no protein is generally recessive, since the gene’s function can be compensated for by the remaining normal copy of the gene.
74 Sons will have a normal phenotype and carry one copy of the normal gene. Daughters will carry one normal gene and one recessive colorblindness allele and will have the normal phenotype.
75 The genotype of the male parent must be XFE Y. The predominance of the normal hair-extra toes and fuzzy hair-normal toes phenotypes in the F1 generation indicates that the female parent must have one X chromosome with both dominant alleles together, and one X chromosome with both recessive alleles together, in other words, her genotype must be XFE Xfe. The fuzzy hair-extra toes and normal hair-normal toes phenotypes are much less common and must be the result of recombination in the female parent, producing XFe and XfE chromosomes. Note that recombination between the X and Y chromosomes in males is not possible due to the fact that the X and Y carry different genes (choices A and C are wrong). If recombination had not occurred in the female parent, all F1 males would have received either XFE or Xfe, giving both normal hair-extra toes and fuzzy hair-normal toes phenotypes (choice B is wrong). Choice D is the correct answer: The F1 females must also have recombinant genotypes on the X chromosomes they received from their mother, but every F1 female also received both dominant alleles on the X chromosome they received from their father. Thus only the dominant phenotypes are seen in the F1 females.
76 Since the affected individuals have unaffected parents, the disease is most likely recessive (choice A is wrong), and since the affected individuals are all male, it is most likely sex-linked (choice B is wrong). There is no father-to-son transmission (all affected males have an unaffected father), so the disease is X-linked (choice C is correct and choice D is wrong).
77 The disease is X-linked recessive, and IIe (IIIa’s father) is not affected, so IIe cannot pass the allele on. Thus the probability of IIIa getting the disease depends on the genotype of IId (she would have to be a carrier) and what she passes on to IIIa. It also depends on the gender of IIIa; females would not be affected because they would have to receive the allele from both IId and IIe, and IIe does not carry the disease allele. Bottom line, in order for IIIa to get the disease, IId would have to be a carrier, would have to pass the disease allele on, and IIIa would have to be male. The probability of IId being a carrier is 1/2; we know she received a good X chromosome from her father Ia (all he has is a good X chromosome), and the probability she received the affected X chromosome from her mother (Ib) is 1/2. The probability she passes the bad X chromosome on to IIIa is 1/2; she has one good X and one bad X. The probability that IIIa is male is 1/2. Finally, we can use the rule of multiplication to determine the overall probability: 1/2 × 1/2 × 1/2 = 1/8 overall probability (choice B is correct and choices C and D are wrong). Note that choice A is wrong because being male does not guarantee the disease; IIIa could be male (get the Y chromosome from IIe) and still be unaffected (get the good X chromosome from IId).
78 Yes. A population does not need to live with each other, only to reproduce sexually with each other.
79 There are two copies of the gene in each of the 2000 individuals, for a total of 4000 copies in the gene pool.
80 The allele frequency is the number of copies of a specific allele divided by the total number of copies of the gene in the population. If there are 5000 hippos, and each has 2 copies of the gene, there are 10,000 copies of the gene in the population. There are 100 homozygotes of the h allele, each with 2 copies of it, and 400 heterozygotes with one h allele, for a total of 600 h alleles in the population. Thus, the frequency of the h allele is 600/10000 = 0.06.
81 In this case, the number of individuals in the population is not provided, but it is not needed. The total number of alleles is 100%. The frequency of the allele is 0.5 × (20% heterozygotes) + 10% homozygotes = 20%.
82 The yellow alleles are still there (but in the heterozygous state) so they do not appear in the phenotype.
83 The frequency of the yellow allele will be 50%, just as it was in the parents. None of the alleles in a population were destroyed, so the frequency is the same as in the parental generation.
84 According to Hardy-Weinberg, there will be no change in the frequency of the allele. The frequency of the yellow allele will still be 50% after four generations.
85 Yes. Independent assortment is not a requirement of Hardy-Weinberg. Allele frequencies for the genes will still remain constant, regardless of the extent of recombination between the genes, as long as the assumptions of Hardy-Weinberg hold true.
86 Hardy-Weinberg says nothing about this situation. Once the assumptions no longer hold true, Hardy-Weinberg no longer applies.
87 If the frequency of the G allele (p) is 0.25, then the frequency of the g allele (q) must be 0.75, since p + q = 1. The frequency of the heterozygotes in the population will be 2pq = 2(0.25)(0.75) = 0.375. Therefore, the number of individuals in this population who are heterozygotes will be 0.375 × 1000 = 375.
88 Genotype frequencies as well as allele frequencies will remain constant according to Hardy-Weinberg.
89 No. The next generation will include GG, Gg, and gg genotypes.
90 The population was not at Hardy-Weinberg equilibrium to start out.
91 Yes. A population reaches Hardy-Weinberg equilibrium after one generation. The F2 generation (and all generations after that) will have the same genotype frequencies as the F1 generation.
92 The allele will not affect fitness. The bears will only be affected at a time when they can no longer have offspring, so it will not affect the ability of bears to transmit their alleles to future generations.
93 If the allele is truly recessive, it will not affect fitness at all. Natural selection can act only on phenotypes, not genotypes.
94 No. Natural selection only acts on heriTable traits. Infected bone marrow cells will not be passed on in the germ line to the next generation and so the long life span of these mice is not a heriTable trait.
95 The fish will technically have the same fitness, since both will contribute to the gene pool of future generations equally.
96 Natural selection acts on phenotypes, not genotypes. Even if the allele is lethal in homozygotes, heterozygotes will not be selected against if the allele is not expressed. It takes many generations for deleterious recessive alleles to decrease in frequency in a population.
97 The correct answer is C. Homozygotes have low fitness in the absence of medicine, while heterozygotes have increased fitness due to their resistance to viral disease. In the absence of the medicine, natural selection tends to reduce the frequency of the allele by removing individuals who are homozygous but tends to increase the frequency of the allele through the higher fitness of heterozygotes. Over time these opposing selection pressures can be balanced to keep the allele at a relatively constant frequency. If medicine removes the selection against homozygotes, then the heterozygotes with the increased fitness cause to allele frequency to increase over time.
98 If there is only one allele, then there is no variability that natural selection can act on, and no way that allele frequencies can change to cause evolution.
99 No. Natural selection can only alter the frequency of existing alleles, not create new alleles.
100 New alleles caused by mutation generally render gene products less active or even inactive. Animals have adapted over long periods of time to have most gene products function in the optimal manner, so most changes are harmful rather than beneficial.
101 No. Mutation must occur in the germ line to introduce a new allele into a population. A mutation in a somatic cell cannot be passed on to the next generation.
102 No. Mitosis can only copy a cell into an identical cell; it is not involved in creating new combinations of alleles in the same manner as meiosis.
103 If the flowers can only reproduce asexually, then they have lost the ability of meiosis to generate new combinations of alleles and new genetic variation for natural selection to act on.
104 Self-pollination reduces genetic variability. More variability is maintained in the population if different individuals mate, making new combinations of alleles.
105 Nonrandom mating and random drift will alter allele frequencies but do not create new alleles (A and B are incorrect). Recombination will not alter allele frequencies or create new alleles, but create new combinations of alleles (C is wrong). The correct answer is D. Only mutation of the genome can create new alleles. A deletion can create a new allele, even if the new allele is a truncated gene product or does not express any gene product at all.
106 Members of a species can mate and produce fit offspring. Members of a population do. Remember it this way: A population is a subset of a species.
107 No, since their offspring are unfit (unable to reproduce).
108 A mnemonic goes: “Dumb King Philip Came Over From Greece Sunday” (or “Dumb King Phil Came Over For Great …”).
109 Note that the arthropods have an exoskeleton surrounding their entire bodies which is composed of the same chemical comprising the fungal cell wall.
110 The dorsal-ventral axis slices the body in half, separating front from back. The axis of symmetry separates left from right. So the two axes are perpendicular. Remember: The axes are planes, not lines.