CONCEPTS IN BIOLOGY
PART III. MOLECULAR BIOLOGY, CELL DIVISION, AND GENETICS
11. Applications of Biotechnology
11.2. Comparing DNA
It is useful to distinguish between individual organisms on the basis of their DNA. Comparisons of DNA can be accomplished in two general ways. Both rely on the fact that genetically different organisms will have different nucleotide sequences in their DNA. The two methods are DNA fingerprinting and DNA sequencing. DNA fingerprinting looks at patterns in specific portions of the DNA of an organism. DNA sequencing looks directly at the nucleotide sequence.
Because both of these approaches have advantages and disadvantages, scientists choose between them depending on their needs. DNA fingerprinting allows for a relatively quick look at larger areas of the organism’s genetic information. It is useful to distinguish between organisms—such as possible suspects in a court trial. DNA sequencing creates a very detailed look at a relatively small region of the organism’s genetic information. DNA sequencing is the most detailed look that we are able to have of the organism’s genetic information.
DNA Fingerprinting
DNA fingerprinting is a technique that uniquely identifies individuals on the basis of short pieces of DNA. Because no two people have the same nucleotide sequences, they do not generate the same lengths of DNA fragments when their DNA is cut with enzymes. Even looking at the many pieces of DNA that are produced in this manner is too complex. Therefore, scientists don’t look at all the possible fragments but, rather, focus on differences found in pieces of DNA that form repeating patterns in the DNA. By focusing on these regions with repeating nucleotide sequences, it is possible to determine whether samples from two individuals have the same number of repeating segments (Outlooks 11.1).
OUTLOOKS 11.1
The First Use of a DNA Fingerprint in a Criminal Case
In 1988, a baker in England was the first person in the world to be convicted of a crime on the basis of DNA evidence. Colin Pitchfork's crime was the rape and murder of two girls. The first murder occurred in 1983. The initial evidence in this case consisted of the culprit's body fluids, which contained his proteins and DNA. On the basis of the proteins, the police were able to create a molecular description of the culprit. The problem was that this description matched 10% of the males in the local population, and the police were unable to identify just one person. In 1986, there was another murder that closely matched the details of the 1983 killing. Another male, Richard Buckland, was the prime suspect for the second murder. In fact, while being questioned, Buckland admitted to the most recent killing but had no knowledge of the first killing. The clues still did not point consistently to a single person.
Meanwhile, the scientists at a nearby university had been working on a new forensic technique—DNA fingerprinting.
To track down the killer, police asked local men to donate blood or saliva samples. Between 4,000 and 5,000 local men participated in the dragnet. None of the volunteers matched the culprit's DNA. Interestingly, Buckland's DNA did not match the culprit's DNA, either. He was later released because his confession was false. It wasn't until after someone reported that Colin Pitchfork had asked a friend to donate a sample for him and offered to pay several others to do the same that police arrested Pitchfork. Pitchfork's DNA matched that of the killer's.
This is a good example of how biotechnology helps the search for truth within the justice system. The additional evidence from DNA was able to provide key information to identify the culprit.
DNA Fingerprinting Techniques
In the scenario presented in Outlooks 11.1, a crime was committed and the scientists had evidence in the form of body fluids from the criminal. These body fluids contained cells with the criminal’s DNA. The DNA in these cells was used as a template to produce enough DNA for analysis. The polymerase chain reaction (PCR) is a technique used to generate large quantities of DNA from small amounts (How Science Works 11.1).
Using PCR and the suspect’s DNA, scientists were able to replicate regions of human DNA that are known to vary from individual to individual. This created large quantities of DNA so that DNA fingerprinting could be performed. Scientists target areas of the suspect’s DNA that contains variable number tandem repeats. Variable number tandem repeats (VNTRs) are sequences of DNA that are repeated a variable number of times from one individual to another. For example, in a given region of DNA, one person may have a DNA sequence repeated 4 times, whereas another may have the same sequence repeated 20 times (figure 11.1).
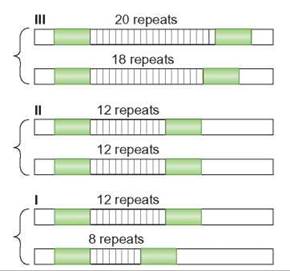
FIGURE 11.1. Variable Number Tandem Repeats
Variable number tandem repeats (VNTRs) are short sequences of DNA that are repeated often. The repeated sequences are attached end-to-end. This illustration shows the VNTRs for three individuals. The individual in I has 8 repeats on 1 chromosome and 12 on the homologous chromosome. They are heterozygous. The individual in II is homozygous for 12 repeats. The individual in III is heterozygous for a different number of repeats—18 and 20.
Once enough DNA was generated through PCR, the DNA needed to be treated so that the VNTRs would be detectable. To detect the varying number of VNTRs, the replicated DNA sample is cut into smaller pieces with restriction enzymes. Restriction sites are DNA nucleotide sequences that attract restriction enzymes. When the restriction enzymes bind to a restriction site, the enzyme cuts the DNA molecule into two molecules. Restriction fragments are the smaller DNA fragments that are generated after the restriction enzyme has cut the selected DNA into smaller pieces. Some of the fragments of DNA that are generated by restriction enzymes will contain the regions with VNTRs. The fragments with VNTRs will vary in size from person to person because some individuals have more repeats than others. Restriction enzymes are used to create fragments of DNA that might be different from one individual to the next.
In DNA fingerprinting, scientists look for different lengths of restriction fragments as an indicator of differences in VNTRs.
Electrophoresis is a technique that separates DNA fragments on the basis of size (How Science Works 11.2). The shorter DNA molecules migrate more quickly than the long molecules. As differently sized molecules are separated, a banding pattern is generated. Each band is a differently sized restriction fragment. Each person’s unique DNA banding pattern is called a DNA fingerprint (figure 11.2). The process of DNA fingerprinting includes the following basic stages:
1. DNA is obtained from a source, which may be as small as one cell.
2. PCR is used to make many copies of portions of the DNA that contain VNTRs.
3. Restriction enzymes are used to cut the VNTR DNA into pieces so that the VNTRs can be detected.
4. To detect the differences in the VNTRs, the pieces are separated by electrophoresis.
5. Comparisons between patterns can be made.
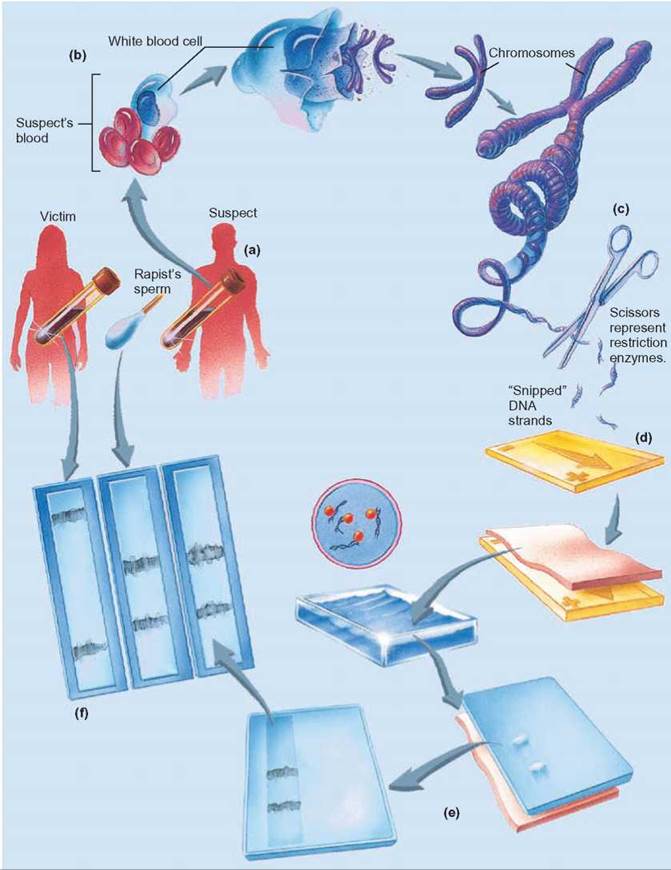
FIGURE 11.2. DNA Fingerprints
(a) Because every person’s DNA is unique, (b) when samples of an individual’s DNA are collected and subjected to restriction enzymes, the cuts occur in different places and DNA fragments of different sizes result. (c) Restriction enzymes can cut DNA at places where specific sequences of nucleotides occur. (d) When the cut DNA fragments are separated by electrophoresis, (e) the smaller fragments migrate more quickly than the larger fragments. This produces a pattern, called a DNA fingerprint, that is unique and identifies the person who provided the DNA. (f) The victim’s DNA is on the left. The rapist’s DNA is in the middle. The suspect’s DNA is on the right. The match in banding patterns between the suspect and the rapist indicates that they are the same.
DNA Fingerprinting Applications
With DNA fingerprinting, the more similar the banding patterns are from two different samples, the more likely the two samples are from the same person. The less similar the patterns, the less likely the two samples are from the same person. In criminal cases, DNA samples from the crime site can be compared with those taken from suspects. If 100% of the banding pattern matches, it is highly probable that the suspect was at the scene of the crime and is the guilty party. The same procedure can be used to confirm a person’s identity, as in cases of amnesia, murder, or accidental death.
DNA fingerprinting can be used in paternity cases that determine the biological father of a child. A child’s DNA is a unique combination of both the mother’s DNA and the father’s DNA. The child’s DNA fingerprint is unique, but all the bands in the child’s DNA fingerprint should be found in either the mother’s or the father’s fingerprint. To determine paternity, the child’s DNA, the mother’s DNA, and DNA from the man who is alleged to be the father are collected.
The DNA from all three is subjected to PCR, restriction enzymes, and electrophoresis. During analysis of the banding patterns, scientists account for the child’s banding pattern by linking each DNA band to a DNA band of the mother and the presumed father. Bands that are common to both the biological mother and the child are identified and eliminated from further consideration. If all the remaining bands can be matched to the presumed father, it is extremely likely that he is the father (figure 11.3). If there are bands that do not match the presumed father’s, then there are one of two conclusions: (1) The presumed father is not the child’s biological father, or (2) the child has a new mutation that accounts for the unique band. This last possibility can usually be ruled out by considering multiple regions of DNA, because it is extremely unlikely that the child will have multiple new mutations.

FIGURE 11.3. Paternity Determination
(a) This illustration shows the VNTRs for four different individuals—a child, the mother, one possible father, and a second possible father. (b) Using PCR, electrophoresis, and DNA fingerprinting analysis, it is possible to identify the child’s father. The mother possesses the “12” band and has passed that to her child. The mother did not give the child the child’s “8” band because the mother does not have an “8” band herself. The child’s “8” band must have come from the father. Of the two men under consideration, only man IV has the “8” band, so man IV is the father. Now stop for a moment and think about the principles of genetics. If man IV is the father, why doesn’t the child have an “18” band?
HOW SCIENCE WORKS 11.1
Polymerase Chain Reaction
Polymerase chain reaction (PCR) is a laboratory procedure for copying selected segments of DNA from larger DNA molecules. With PCR, a single cell can provide enough DNA for analysis and identification. Scientists start with a sample of DNA that contains the desired DNA region. The types of samples that can be used include semen, hair, blood, bacteria, protozoa, viruses, mummified tissues, and frozen cells. Targeting specific portions of DNA for replication enables biochemists to manipulate DNA more easily. When many copies of this DNA have been produced it is easy to find, recognize, and manipulate.
PCR is a test-tube version of the cellular DNA replication process and requires similar components. The DNA from the sample specimen serves as the template for replication. Free DNA nucleotides are used to assemble new strands of DNA. DNA polymerase, which has been purified from bacteria cells, is used to catalyze the PCR reaction.
DNA primers are short stretches of single-stranded DNA, which are used to direct the DNA polymerase to replicate only certain regions of the template DNA. These primer molecules are specifically designed to flank the ends of the target region's DNA sequence and point the DNA polymerase to the region between the primers. The PCR reaction is carried out by heating the target DNA, so that the two strands of DNA fall away from each other. This process is called denaturation. Once the nitrogenous bases on the target sequence are exposed and the reaction cools, the primers are able to attach to the template molecule. The primers anneal to the template. The primers anneal (that is, stick or attach) to the template. The primers are able to target a particular area of DNA because the primer nucleotide sequence pairs with the template DNA sequence using the base-pairing rules.
Purified DNA polymerase is the enzyme that drives the DNA replication process. The presence of the primer, attached to the DNA template and added nucleotides, serves as the substrate for the DNA polymerase. Once added, the polymerase extends the DNA molecule from the primer down the length of the DNA. Extension continues until the polymerase falls off of the template DNA. The enzyme incorporates the new DNA nucleotides in the growing DNA strand. It stops when it reaches the other end, having produced a new copy of the target sequence.
The elegance of PCR is that it allows the exponential replication of DNA. Exponential, or logarithmic, growth is a doubling in number with each round of PCR. With just one copy of template DNA, there will be a total of two copies at the end of one replication cycle. During the second round, both copies are used as a template. At the end of the second round, there is a total of 4 copies. The number of copies of the target DNA increases very quickly. With each round of replication, the number doubles—8, 16, 32, 64. Each round of replication takes only minutes. Thirty rounds of replication in PCR can be performed within 2.5 hours. Starting with just one copy of DNA and 30 rounds of replication, it is possible to produce over half a billion copies of the desired DNA segment.
Because this technique can create useful amounts of DNA from very limited amounts, it is a very sensitive test for the presence of specific DNA sequences. Frequently, the presence of a DNA sequence indicates the presence of an infectious agent or a disease-causing condition.
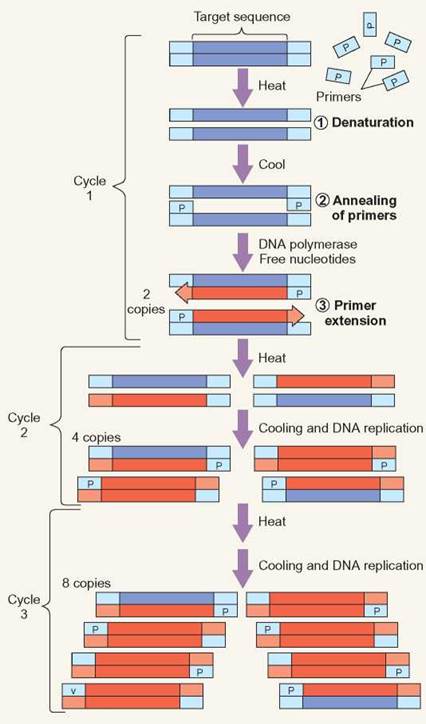
PCR Replication
During cycle one of PCR, the template DNA is denatured, so that the two strands of DNA separate. This allows the primers to attach (anneal) to the template DNA. DNA polymerase and DNA nucleotides, which are present for the reaction, create DNA by extending from the primers. During cycle 2, the same process occurs again, but the previous round of replication has made more template available for further replication. Each subsequent cycle essentially doubles the amount of DNA.
HOW SCIENCE WORKS 11.2
Electrophoresis
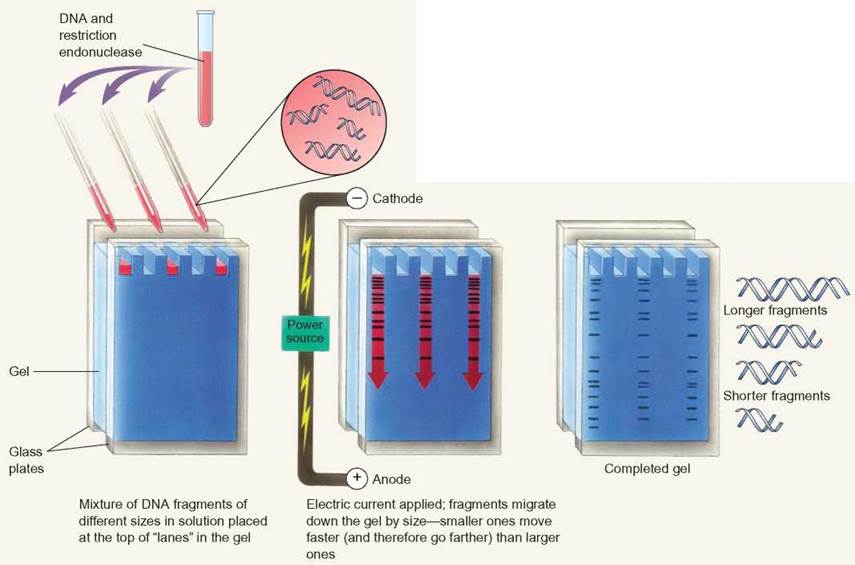
Electrophoresis is a technique used to separate molecules, such as nucleic acids, proteins, or carbohydrates. Electrophoresis separates nucleic acids on the basis of size. DNA is too long for scientists to work with when taken directly from the cell. To make the DNA more manageable, scientists cut the DNA into smaller pieces. Restriction enzymes are frequently used to cut large DNA molecules into smaller pieces. After the DNA is broken into smaller pieces, electrophoresis is used to separate differently sized DNA fragments.
Electrophoresis uses an electric current to move DNA through a gel matrix. DNA has a negative charge because of the phosphates that link the nucleotides. In an electrical field, DNA migrates toward the positive pole. The speed at which DNA moves through the gel depends on the length of the DNA molecule. Longer DNA molecules move more slowly through the gel matrix than do shorter DNA molecules.
When scientists work with small areas of DNA, electrophoresis allows them to isolate specific stretches of DNA for other applications.
HOW SCIENCE WORKS 11.3
DNA Sequencing
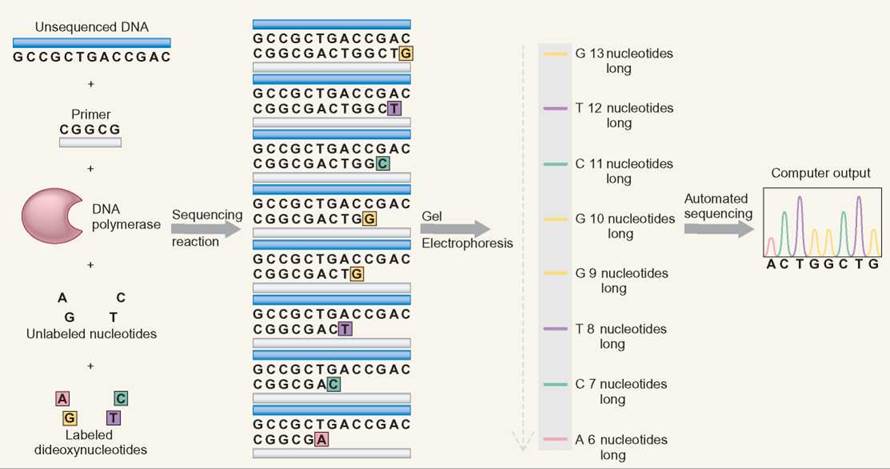
DNA sequencing uses electrophoresis to separate DNA fragments of different lengths. A DNA synthesis reaction is set up that includes DNA from the region being investigated. The reaction also includes (1) DNA polymerase, (2) a specific DNA primer, (3) all DNA nucleotides (G, A, T, and C), and (4) a small amount of 4 kinds of chemically altered DNA nucleotides. DNA polymerase is the enzyme that synthesizes DNA in cells by using DNA nucleotides as a substrate. The DNA primer gives the DNA polymerase a single place to start the DNA synthesis reaction. All of these components work together to allow DNA synthesis in a manner very similar to cellular DNA replication.
The DNA sequencing process also adds nucleotides that have been chemically altered in two ways: (1) The altered nucleotides are called dideoxyribonucleosides because they contain a dideoxyribose sugar rather than the normal deoxyribose. Dideoxyribose has one less oxygen in its structure than deoxyribose. (2) The four kinds of dideoxyribonucleotides (A, T, G, C) are each labeled with a different flourescent dye so that each of the four nucleotides is colored differently. During DNA sequencing, the DNA polymerase randomly incorporates either a normal DNA nucleotide or a dideoxyribonucleotide. When the dideoxyribonucleoside is used, two things happen: (1) No more nucleotides can be added to the DNA strand, and (2) the DNA strand is now tagged with the fluorescent label of the dideoxynucleotide that was just incorporated.
As a group, the DNA molecules that are created by this technique have the following properties:
1. They all start at the same point, because they all started with the same primer.
2. There are copies of DNA molecules that had their replication halted at each nucleotide in the sequence of the sample DNA when a dideoxyribose nucleotide was incorporated.
3. DNA molecules of the same length (number of nucleotides) are labeled with the same color of fluorescent dye.
Electrophoresis separates this collection of molecules by size, because the shortest DNA molecules move fastest. The DNA sequence is determined by reading the color sequence from the shortest DNA molecules to the longest DNA molecules. The pattern of colors matches the order of the nucleotides in the DNA. The color pattern that is generated by the sequencing gel is the order of the nucleotides. Automated sequencing is done by using a laser beam to read the colored bands. A printout is provided as peaks of color to show the order of the nucleotides.
Gene Sequencing and the Human Genome Project
The Human Genome Project (HGP) was a 13-year effort to determine the human DNA sequence. Work began in 1990. It was first proposed in 1986 by the U.S. Department of Energy (DOE) and was cosponsored soon after by the National Institutes of Health (NIH). These agencies were the main research agencies within the U.S. government responsible for developing and planning the project. Estimates are that the United States spent over $3 billion on the Human Genome Project.
Many countries contributed both funds and labor resources to the Human Genome Project. At least 17 countries other than the United States participated, including Australia, Brazil, Canada, China, Denmark, France, Germany, Israel, Italy, Japan, Korea, Mexico, the Netherlands, Russia, Sweden, and the United Kingdom. The Human Genome Project was one of the most ambitious projects ever undertaken in the biological sciences.
The data that these countries produced are stored in powerful computers, so that the information can be shared. To get an idea of the size of this project, consider that a human Y chromosome (one of the smallest of the human chromosomes) is composed of nearly 60 million paired nucleotides. The larger X chromosome may be composed of 150 million paired nucleotides. The entire human genome consists of 3.12 billion paired nucleotides. That is roughly the same number as all the letter characters found in about 2,000 copies of this textbook.
Human Genome Project Techniques
Two kinds of work progressed simultaneously to determine the sequence of the human genome. First, physical maps were constructed by determining the location of specific “markers” and the proximity of these markers to genes. The markers were known sequences of DNA that could be located on the chromosome. This physical map was used to organize the vast amount of data produced by the second technique, which was for the labs to determine the exact order of nitrogenous bases of the DNA for each chromosome. Techniques exist for determining base sequences (How Science Works 11.3). The challenge is storing and organizing the information from these experiments, so that the data can be used.
A slightly different approach was adopted by Celera Genomics, a private U.S. corporation. Although Celera Genomics started later than the labs funded by the Department of Energy and National Institute of Health it was able to catch up and completed its sequencing at almost the same time as the government-sponsored programs by developing new techniques. Celera jumped directly to determining the DNA sequence of small pieces of DNA without the physical map. It then used computers to compare and contrast the short sequences, so that it could put them together and assemble the longer sequence. The benefit of having these two organizations as competitors was that, when they finished their research, they could compare and contrast results. Amazingly, the discrepancies between their findings were declared insignificant.
Human Genome Project Applications
The first draft of the human genome was completed early in 2003, when the complete nucleotide sequence of all 23 pairs of human chromosomes was determined. By sequencing the human genome, it is as if we have now identified all the words in the human gene “dictionary.” Continued analysis will provide the definitions for these words—what these words tell the cell to do.
The information provided by the human genome project is extremely useful in diagnosing diseases and providing genetic counseling to those considering having children. This information can identify human genes and proteins that can be targets for drugs and new gene therapies. Once it is known where an abnormal gene is located and how it differs in base sequence from the normal DNA sequence, steps could be taken to correct the abnormality. Further defining the human genome will also result in the discovery of new families of proteins and will help explain basic physiological and cell biological processes common to many organisms. All this information will increase the breadth and depth of the understanding of basic biology.
It was originally estimated that there were between 100,000 and 140,000 genes in the human genome, because scientists were able to detect so many different proteins. DNA sequencing data indicate that there are only about 20,000 protein-coding genes—only about twice as many as in a worm or a fly. Our genes are able to generate several different proteins per gene because of alternative splicing (figure 11.4). Alternative splicing occurs much more frequently than previously expected. Knowing this information provides insights into the evolution of humans and will make future efforts to work with the genome through bioengineering much easier.
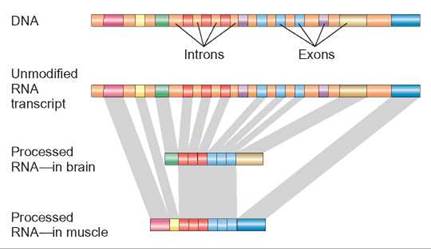
FIGURE 11.4. Different Proteins—One Gene
This illustration shows a stretch of DNA that contains a gene. Proteincoding regions (exons) of this gene are shown in different colors. Introns that do not code for protein and are not transcribed into RNA are shown in a single color—rust. Alternative splicing allows different tissue to use the same gene but make slightly different proteins. The gray bands show how some exons are used to form both proteins, whereas other exons are used on only one protein.
There is a concern that, as our genetic makeup becomes easier to determine, some people may attempt to use this information for profit or political power. Consider that some health insurance companies refuse to insure people with “preexisting conditions” or those at “genetic risk” for certain abnormalities. Refusing to provide coverage would save these companies the expense of future medical bills incurred by “less than perfect” people. While this might be good for insurance companies, it raises major social questions about fair and equal treatment and discrimination.
Another fear is that attempts may be made to “breed out” certain genes and people from the human population to create a “perfect race.” Intentions such as these superficially appear to have good intentions, but historically they have been used by many groups to justify discrimination against groups of individuals or even to commit genocide.
Other Genomes
While some scientists refine our understanding of the human genome, others are sequencing the genomes of other organisms. Representatives of each major grouping of organisms have been investigated, and the DNA sequence data have been made available to the general public through a centralized government website. This centralized database has made the exchange and analysis of scientific information easier than ever (table 11.1). The information gained from these studies
TABLE 11.1. Completed and Current Genome Projects
The Human Genome Project has sparked major interest in nonhuman genomes. The investigation of some genomes has been very organized. Other investigations have been less directed, whereby only sequences of certain regions of interest have been reported. Regardless, information on many genomes is available at the National Center for Biotechnology Information website.
Taxonomic Group |
Genome Examples |
Number of Different Genomes Represented |
Viruses and retroviruses and bacteriphages |
Herpes virus, human papillomavirus, HIV |
Over 1,560 |
Bacteria |
Anthrax species, Chlamydia species, Escherichia coli, Pseudomonas species, Salmonella species |
Over 200 |
Archaea |
Halobacterium species, Methanococcus species, Pyrococcus species, Thermococcus species |
Over 21 |
Protists |
Cryptosporidium species, Entamoeba histolytica, Plasmodium species |
Over 45 |
Fungi |
Yeast, Aspergillus, Candida |
Over 70 |
Plants |
Thale cress (Arabidopsis thaliana), tomato, lotus, rice |
Over 20 |
Animals |
Bee, cat, chicken, chimp, cow, dog, frog, fruit fly, mosquito, nematode, pig, rat, sea urchin, sheep, zebra fish |
Over 100 |
Cellular organelles |
Mitochondria, chloroplasts |
|
Patterns in Protein-Coding Sequences
As scientists sequenced the human genome and compared it with other genomes, certain patterns became apparent.
Tandem clusters are grouped copies of the same gene that are found on the same chromosome (figure 11.5). For example, the DNA that codes for ribosomal RNA is present in many copies in the human genome. From an evolutionary perspective, the advantage to the cell is the ability to create large amounts of gene product quickly from the genes found in tandem clusters.
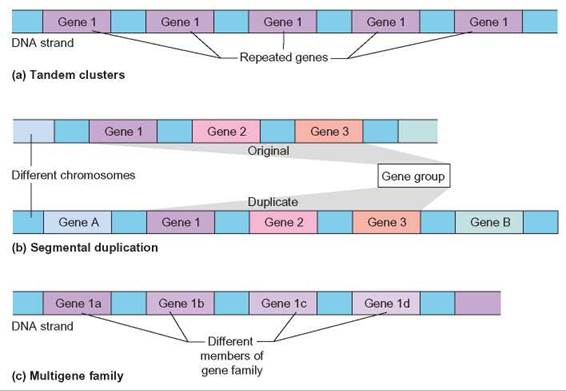
FIGURE 11.5. Patterns in Protein Coding Sequences
(a) Tandem clusters are identical or nearly identical repeats of one gene. (b) Segmental duplications are duplications of sets of genes. These may occur on the same chromosome or different chromosomes. (c) Multigene families are repeats of similar genes. The genes are similar because regions are conserved from gene to gene, but many regions have changed significantly.
Segmental duplications are groups of genes that are copied from 1 chromosome and moved as a set to another chromosome. These types of gene duplications allow for genetic backups of information. If either copy is mutated, the remaining copy can still provide the necessary gene product sufficient for the organism to live. Because the function of the mutated copy of the gene is being carried out by the normal gene, the mutated copy may take on new function if it accumulates additional mutations. This can allow evolution to occur more quickly (figure 11.5b).
Multigene families are groups of different genes that are closely related. When members of multigene families are closely inspected, it is clear that certain regions of the genes carry similar nucleotide sequences. Hemoglobin is a member of the globin gene family. There are several different hemoglobin genes in the human genome. Evolutionary patterns can be tracked at the molecular level by examining gene families across species. The portions of genes that show very little change across many species represent portions of the protein that are important for function. Scientists reason that regions that are important for function will be intolerant of change and stay unaltered over time. Again, using hemoglobin as an example, it is possible to compare the hemoglobin genes of different organisms to identify specific changes in the gene. Such comparisons can lead to a better understanding of how organisms are related to each other evolutionarily (figure 11.5c).
The following are a few more interesting facts obtained by comparing genomes:
• Eukaryotic genomes are more complex than prokaryotic genomes. Eukaryotic genomes are, on average, nearly twice the size of prokaryotic genomes. Eukaryotic genomes devote more DNA to regulating gene expression. Only 1% of human DNA actually codes for protein.
• The number of genes in a genome is not a reflection of the size or complexity of an organism. Humans possess roughly 21,000 genes. Roundworms have about 26,000 genes, and rice plants possess 32,000 to 55,000 genes.
• Eukaryotes create multiple proteins from their genes because of alternative splicing. Prokaryotes do not. Nearly 25% of human DNA consists of intron sequences, which are removed during splicing. On average, each human gene makes between 4.5 and 5 different proteins because of alternative splicing.
• There are numerous, virtually identical genes found in very distantly related organisms—for example, mice, humans, and yeasts.
• Hundreds of genes found in humans and other eukaryotic organisms appear to have resulted from the transfer of genes from bacteria to eukaryotes at some point in eukaryotic evolution.
• Chimpanzees have 98-99% of the same DNA sequence as humans. All the human “races” are about 99.9% identical at the DNA level. In fact, there is virtually no scientific reason for the concept of “race,” because the amount of variation within a race is as great as the amount of variation between races.
• Genes are unequally distributed between chromosomes and unequally distributed along the length of a chromosome.
Patterns in Non-Coding Sequence
Protein-coding DNA is not the only reason for examining DNA sequences. The regions of DNA that do not code for protein are more important than once thought. Many noncoding sequences are involved with the regulation of gene expression.
A recent and more accurate map of the human genome from Britain focuses on copy number variations, or CNVs. These are segments in the genetic code that can be deleted or copied; most are deletions and a small number are duplications. The new map has also revealed that humans have:
1. 75 “jumping genes,” or transposable elements (regions of the genetic code that can move from one place to another in the genome of an individual).
2. more than 250 genes that can lose one of the two copies in chromosomes and not causing any obvious consequences, and
3. 56 genes that can join together, potentially forming new genes.
New Fields of Knowledge
The ability to make comparisons of the DNA of organisms has led to the development of three new fields in biology— genomics, transcriptomics, and proteomics. Genomics is the comparison of the genomes of different organisms to identify similarities and differences. Species relatedness and gene similarities can be determined from these studies. When the DNA sequence of a gene is known, transcriptomics looks at when, where, and how much mRNA is expressed from a gene. Finally, proteomics examines the proteins that are predicted from the DNA sequence. From these types of studies, scientists are able to identify gene families that can be used to determine how humans have evolved at a molecular level. They can also examine how genes are used in an organism throughout its body and over its life span. They can also better understand how a protein works by identifying common themes from one protein to the next.
11.2. CONCEPT REVIEW
3. What types of questions can be answered by comparing the DNA of two different organisms?
4. What techniques do scientists use to compare DNA?
5. What benefits does the Human Genome Project offer?
6. What is the purpose of the PCR?
7. What role does electrophoresis play in DNA comparisons?
8. What are tandem clusters, segmental duplications and multigene families?