Biology of Humans
21. DNA and Biotechnology
In the previous chapter, we learned about the chromosomal basis of inheritance. In this chapter, we become familiar with the structure and function of DNA and discover how this molecule is able to serve as the basis of our genetic inheritance as well as the source for the diversity of life on Earth. We learn that the importance of DNA on a personal level is that it directs the synthesis of specific polypeptides (proteins) that play structural or functional roles in our bodies. We then consider the technology that our understanding of DNA has already made available and what possibilities such technology may hold for the future.
Form of DNA
DNA is sometimes referred to as the thread of life—and a very slender thread it is. When DNA is unwound, it measures a mere 50-trillionths of an inch in diameter. If all the DNA strands in a single cell were fastened together end to end, the thread would stretch more than 5 ft in length. DNA might also be considered the thread that ties all life together, because the DNA of organisms ranging from bacteria to humans is built from the same kinds of subunits. The order of these subunits encodes the information needed to make the proteins that build and maintain life.
Deoxyribonucleic acid, or DNA, is a double-stranded molecule resembling a ladder that is gently twisted to form a spiral called a double helix, as shown in Figure 21.1. Each side of the ladder, including half of each rung, is made from a string of repeating subunits called nucleotides. You may recall from Chapter 2 that a nucleotide is composed of three subunits, including one sugar (deoxyribose, in DNA), one phosphate, and one nitrogenous base. DNA contains four types of nitrogenous bases: adenine (A), guanine (G), thymine (T), and cytosine (C). The sides of the ladder are composed of alternating sugars and phosphates; the rungs consist of paired nitrogenous bases. The bases attach to each other according to the rules of complementary base pairing: adenine pairs only with thymine (creating an A-T pair), and cytosine pairs only with guanine (creating a C-G pair). Each base pair is held together by weak hydrogen bonds. The pairing of complementary bases is specific due to the shapes of the bases and the number of hydrogen bonds that can form between them. You may also recall from Chapter 2 that a molecule formed by the joining of nucleotides is called a nucleic acid. Thus, DNA is a nucleic acid.
· A DNA fingerprint is based on the sequence of bases in a person's DNA. Thus, a DNA fingerprint is unique to each person (except for those who happen to have identical siblings).
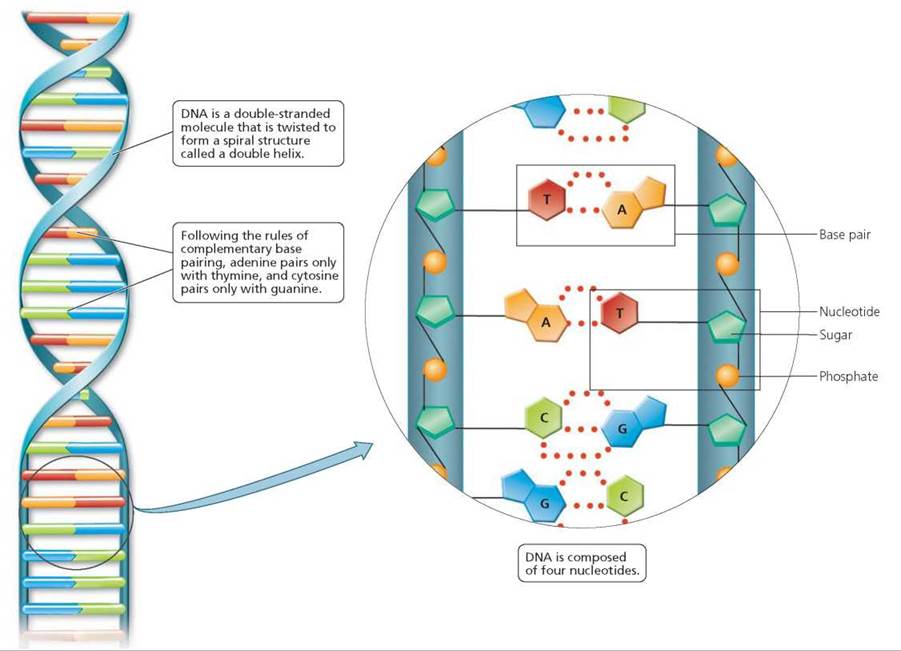
FIGURE 21.1. DNA is a double-stranded molecule that twists to form a spiral structure called a double helix.
Because base pairing is so specific, the bases on one strand of DNA are always complementary to the bases on the other strand. Thus the order of bases on one strand determines the sequence of bases on the other strand. For instance, if the sequence of bases on one strand were CATATGAG, what would the complementary sequence be? Remember, cytosine (C) always pairs with guanine (G), and adenine (A) always pairs with thymine (T). As a result, the complementary sequence on the opposite strand would be GTATACTC.
The DNA within each human cell has an astounding 3 billion base pairs. Although the pairing of adenine with thymine and cytosine with guanine is specific and does not vary, the sequence of bases throughout the length of different DNA molecules can vary in myriad ways. As we will see, genetic information is encoded in the exact sequence of bases.
Replication of DNA
For DNA to be the basis of inheritance, its genetic instructions must be passed from one generation to the next. Moreover, for DNA to direct the activities of each cell, its instructions must be present in every cell. These requirements dictate that DNA be copied before both mitotic and meiotic cell division (see Chapter 19). It is important that the copies be exact. The key to the precision of the copying process is that the bases are complementary.
The copying process, or DNA replication, begins when an enzyme breaks the weak hydrogen bonds that hold together the paired bases that make up nucleotide strands of the double helix, thereby "unzipping" and unwinding the strands. As a result, the nitrogenous bases on the separated regions of each strand are temporarily exposed. Free nucleotide bases, which are always present within the nucleus, can then attach to complementary bases on the open DNA strands. Enzymes called DNA polymerases link the sugars and phosphates of the newly attached nucleotides to form a new strand. As each of the new double-stranded DNA molecules forms, it twists into a double helix.
Each strand of the original DNA molecule serves as a template for the formation of a new strand. This process is called semiconservative replication, because in each of the new double-stranded DNA molecules, one original (parent) strand is saved (conserved) and the other (daughter) strand is new. Look at Figure 21.2. Notice the original (parental) strand of nucleotides in each new molecule of DNA. Complementary base pairing creates two new DNA molecules that are identical to the parent molecule.
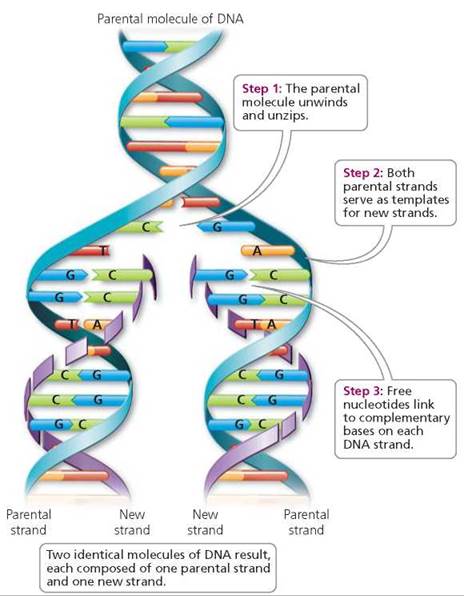
FIGURE 21.2. DNA replication is called semiconservative because each daughter molecule consists of one “parental" strand and one “new" strand.
Gene Expression
The process of replication ensures that genetic information is passed accurately from a parent cell to daughter cells and from generation to generation. The next obvious question is, "How does DNA issue commands that direct cellular activities?" The answer is that DNA directs the synthesis of another nucleic acid— ribonucleic acid, or RNA. RNA, in turn, directs the synthesis of a polypeptide (a part of a protein) or a protein. The protein may be a structural part of the cell or play a functional role, such as an enzyme that speeds up certain chemical reactions within the cell.
Recall from Chapter 20 that a gene is a segment of DNA that contains the instructions for producing a specific protein (or in some cases, a specific polypeptide).1 The sequence of bases in DNA determines the sequence of bases in RNA, which in turn determines the sequence of amino acids of a protein. We say that the gene is expressed when the protein it codes for is produced. The resulting protein is the molecular basis of the inherited trait; it determines the phenotype.
![]()
To more fully appreciate how gene expression works, we will consider each step in slightly greater detail.
RNA Synthesis
Just as the CEO of a major company issues commands from headquarters instead of from the factory floor, DNA issues instructions from the cell nucleus and not from the cytoplasm where the cell's work is done. RNA is the intermediary that carries the information encoded in DNA from the nucleus to the cytoplasm and directs the synthesis of the specified protein.
Like DNA, RNA is composed of nucleotides linked together, but there are some important differences between DNA and RNA, as shown in Table 21.1. First, the nucleotides of RNA contain the sugar ribose, instead of the deoxyribose found in DNA. Second, in RNA the nucleotide uracil (U) pairs with adenine, whereas in DNA thymine (T) pairs with adenine (A). Third, most RNA is single stranded. Recall that DNA is a double-stranded molecule.
TABLE 21.1. Comparisons of DNA and RNA
|
DNA |
RNA |
Similarities |
Are nucleic acids |
|
|
Are composed of linked nucleotides |
|
|
Have a sugar-phosphate backbone |
|
|
Have four types of bases |
|
Differences |
Is a double-stranded molecule |
Is a single-stranded molecule |
|
Has a sugar deoxyribose |
Has a sugar ribose |
|
Contains the bases adenine, guanine, cytosine, and thymine |
Contains the bases adenine, guanine, cytosine, and uracil (instead of thymine) |
|
Functions primarily in the nucleus |
Functions primarily in the cytoplasm |
The first step in converting the DNA message to a protein is to copy the message as RNA, by a process called transcription.
![]()
Three types of RNA are produced in cells. Each plays a different role in protein synthesis (Table 21.2). Messenger RNA (mRNA) carries DNA's instructions for synthesizing a particular protein from the nucleus to the cytoplasm. The order of bases in mRNA specifies the sequence of amino acids in the resulting protein, as we will see. Each transfer RNA (tRNA) molecule is specialized to bring a specific amino acid to where it can be added to a polypeptide that is under construction. Ribosomal RNA (rRNA) combines with proteins to form ribosomes, which are the structures on which protein synthesis occurs.
TABLE 21.2. Review of the Functions of RNA
Molecule |
Functions |
Messenger RNA (mRNA) |
Carries DNA's information in the sequence of its bases (codons) from the nucleus to the cytoplasm |
Transfer RNA (tRNA) |
Binds to a specific amino acid and transports it to be added, as appropriate, to a growing polypeptide chain |
Ribosomal RNA (rRNA) |
Combines with protein to form ribosomes (structures on which polypeptides are synthesized) |
Transcription begins with the unwinding and unzipping of the specific region of DNA to be copied; these actions are performed by an enzyme. The DNA message is determined by the order of bases in the unzipped region of the DNA molecule. One of the unwound strands of the DNA molecule serves as the template during transcription. RNA nucleotides present in the nucleus pair with their complementary bases on the template—cytosine with guanine and uracil with adenine (Figure 21.3). The signal to start transcription is given by a specific sequence of bases on DNA, called the promoter. An enzyme called RNA polymerase binds with the promoter on DNA and then moves along the DNA strand, opening up the DNA helix in front of it and then aligning the appropriate RNA nucleotides and linking them together; the region of DNA that has been transcribed zips again after RNA polymerase passes by. Another sequence of bases on the DNA signals RNA polymerase to stop transcription. After transcription ceases, the newly formed strand of RNA, called the RNA transcript, is released from the DNA.
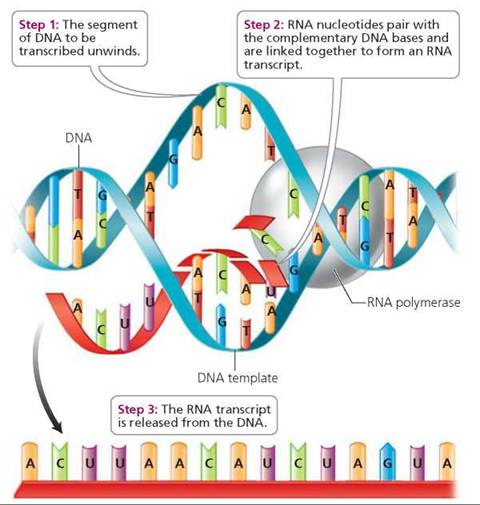
FIGURE 21.3. Transcription is the process of producing RNA from a DNA template.
Messenger RNA usually undergoes certain modifications before it leaves the nucleus (Figure 21.4). Most stretches of DNA between a promoter and the stop signal include regions that do not contain codes that will be translated into protein. These unexpressed regions of DNA are called introns, short for intervening sequences. The regions of mRNA corresponding to the introns are snipped out of the newly formed mRNA strand by enzymes before the strand leaves the nucleus. The remaining segments of DNA or mRNA, called exons for expressed sequences, splice together to form the sequence that directs the synthesis of a protein.
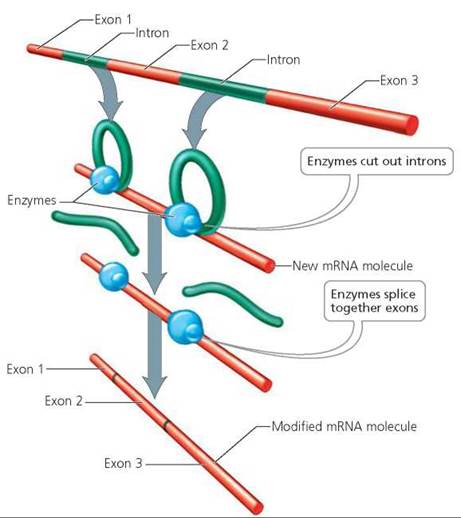
FIGURE 21.4. Newly formed messenger RNA is modified before it leaves the nucleus. Noncoding regions of DNA called introns are snipped out of the corresponding regions of mRNA molecule. Segments of mRNA that code for protein are then spliced together.
Protein Synthesis
The newly formed mRNA carries the genetic message (transcribed from DNA) from the nucleus to the cytoplasm, where it is translated into protein at the ribosomes. Just as we might translate a message written in Spanish into English, translation converts the nucleotide language of mRNA into the amino acid language of a protein.
![]()
Before examining the process of translation, we should become more familiar with the language of mRNA.
The genetic code. To use any language, you must know what the words are and what they mean, as well as where sentences begin and end. The genetic code is the "language" of genes that translates the sequence of bases in DNA into the specific sequence of amino acids in a protein. We have seen that the sequence of bases in DNA determines the sequence of bases in mRNA through complementary base pairing. The "words" in the genetic code, called codons, are sequences of three bases on mRNA that specify 1 of the 20 amino acids or the beginning or end of the protein chain. All the codons of the genetic code are shown in Figure 21.5. For instance, the codon UUC on mRNA specifies the amino acid phenylalanine. (The complementary sequence on DNA would be AAG.)
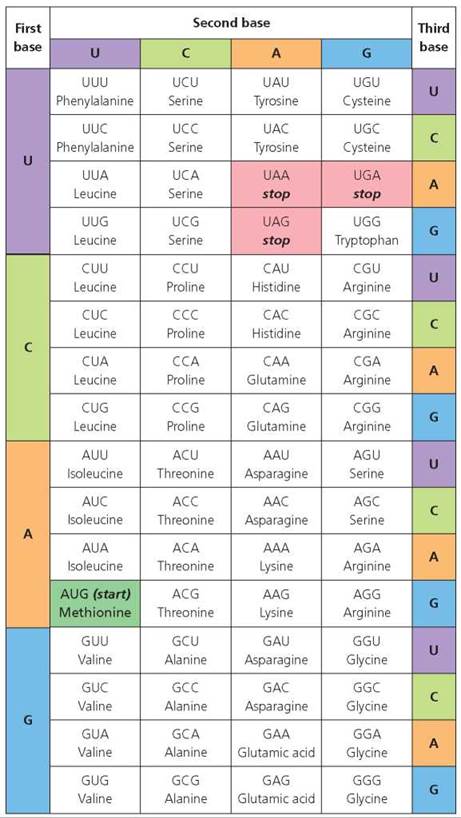
FIGURE 21.5. The genetic code. Each sequence of three bases on the mRNA molecules, called a codon, specifies a specific amino acid, a start signal, or a stop signal.
If the sequence of bases following a start signal were AACUCAGCC, what amino acids would be specified?
Asparagine, serine, alanine
Stop and think
Look at the strand of mRNA in Figure 21.3. Notice that the codon at the end of the mRNA strand is ACG. Which amino acid does this specify? (Use Figure 21.5.)
The four bases in RNA (A, U, C, and G) could form 64 combinations of three-base sequences. The number of possible codons, therefore, exceeds the number of amino acids. As Figure 21.5 indicates, there are several sets of codons that code for the same amino acid. Note, too, that the codon AUG can either serve as a start signal to initiate translation or can specify the addition of the amino acid methionine to the growing protein chain, depending on where it occurs in the mRNA molecule. In addition, three codons (UAA, UAG, and UGA) are stop codons that signal the end of a protein and that do not code for an amino acid. If we think of the codons as genetic words, then a stop codon functions as the period at the end of the sentence.
Transfer RNA A language interpreter translates a message from one language to another. Transfer RNA (tRNA) serves as an interpreter that converts the genetic message carried by mRNA into the language of protein, which is a particular sequence of amino acids. To accomplish this conversion, a tRNA molecule must be able to recognize both the codon on mRNA and the amino acid that the codon specifies—in other words, it must speak both languages.
There are many kinds of tRNA—at least one for each of the 20 amino acids. Each type of tRNA molecule binds to a particular amino acid. Enzymes ensure tRNA binds with the correct amino acid. The tRNA then ferries the amino acid to the correct location along a strand of mRNA (Figure 21.6).
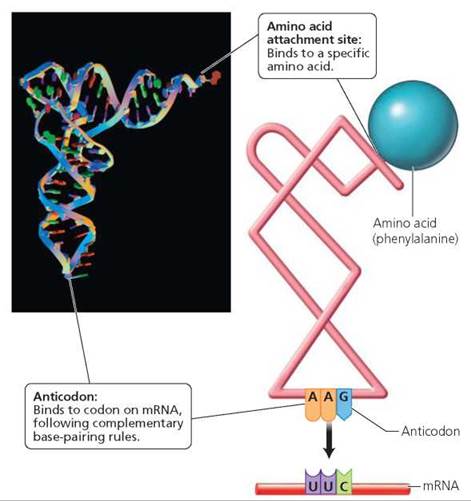
FIGURE 21.6. A tRNA molecule Is a short strand of RNA that twists and folds on Itself. The job of tRNA Is to ferry a specific amino acid to the ribosome and insert it in the appropriate position in the growing peptide chain.
How does the tRNA know the correct location along mRNA? The location is determined by a sequence of three nucleotides on the tRNA called the anticodon. In a sense, the anticodon "reads" the language of mRNA by binding to a codon on the mRNA molecule according to the complementary base-pairing rules. When the tRNA's anticodon binds to the mRNA's codon, the specific amino acid attached to the tRNA is brought to the growing polypeptide chain. For example, a tRNA molecule with the anticodon AAG binds to the amino acid phenylalanine and ferries it to the mRNA molecule, where the codon UUC is presented for translation. Phenylalanine will then be added to the growing amino acid chain.
Ribosomes. Ribosomes function as the workbenches on which proteins are built from amino acids. A ribosome consists of two subunits (small and large), each composed of ribosomal RNA (rRNA) and protein. The subunits form in the nucleus and are shipped to the cytoplasm. They remain separate except during protein synthesis. The role of the ribosome in protein synthesis is to bring the tRNA bearing an amino acid close enough to the mRNA to interact. As you can see in Figure 21.7, when the two subunits fit together to form a functional ribosome, a groove for mRNA is formed. Two binding sites position tRNA molecules so that an enzyme in the ribosome can cause bonds to form between their amino acids.
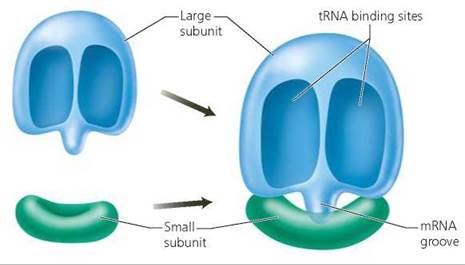
FIGURE 21.7. A ribosome consists of two subunits of different sizes. When the two subunits join together to form a functional ribosome, a groove for mRNA is formed. The ribosome has two binding sites for tRNA molecules. It also contains an enzyme that promotes the formation of a peptide bond between the amino acids that are attached to the tRNAs in the binding sites.
Protein synthesis. Translation—essentially, protein synthesis— can be divided into three stages: initiation, elongation, and termination.
1. During initiation, the major players in protein synthesis (mRNA, tRNA, and ribosomes) come together (Figure 21.8).
• Step 1: The small ribosomal subunit attaches to the mRNA strand at the start codon, AUG.
• Step 2: The tRNA with the complementary anticodon pairs with the start codon. The larger ribosomal subunit then joins the smaller one to form a functional, intact ribosome with mRNA positioned in a groove between the two subunits.
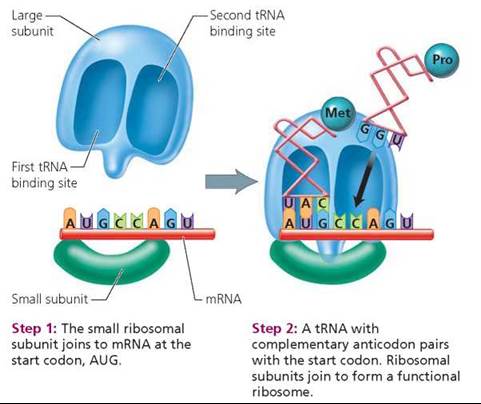
FIGURE 21.8. Initiation of translation
2. Elongation of the protein occurs as additional amino acids are added to the chain (Figure 21.9).
• Step 1: Codon recognition. With the start codon positioned in one binding site, the next codon is aligned in the other binding site.
• Step 2: Peptide bond formation. The tRNA bearing an anticodon that will pair with the exposed codon slips into place at the binding site, and the amino acid it carries forms a peptide bond with the previous amino acid with the assistance of enzymes.
• Step 3: Ribosome movement. The tRNA in the first binding site leaves the ribosome. The ribosome moves along the mRNA molecule, carrying the growing peptide chain and the remaining tRNA with its amino acid to the first binding site. This movement positions the next codon in the open site. An appropriate tRNA slips into the open site, and its amino acid binds to the previous one. This process is repeated many times, adding one amino acid at a time to the growing polypeptide chain.
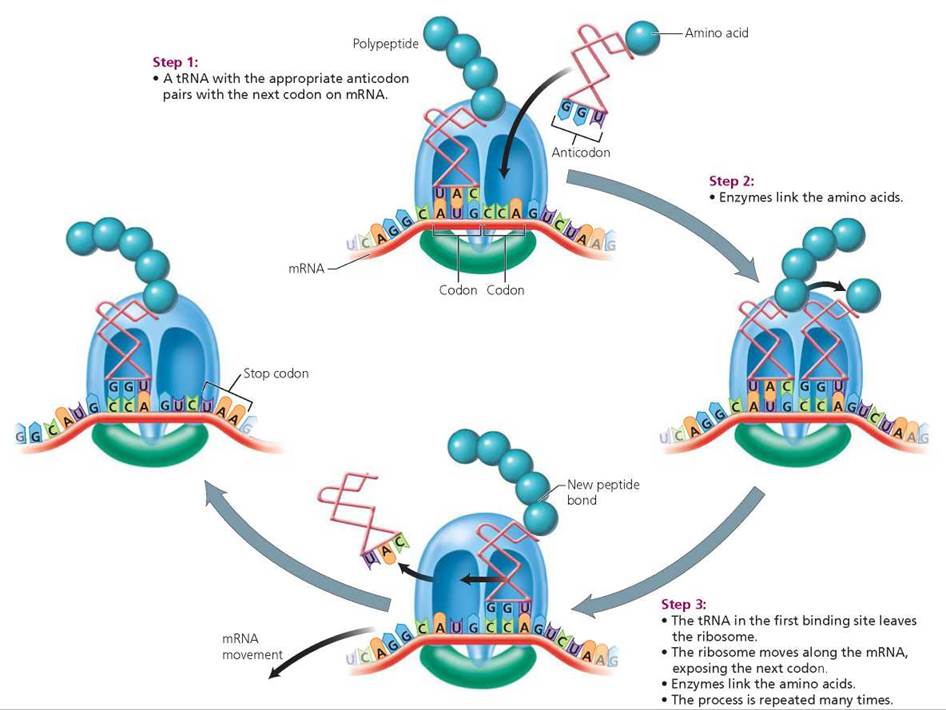
FIGURE 21.9. Elongation of the polypeptide during translation
Many ribosomes may glide along a given mRNA strand at the same time, each producing its own copy of the protein directed by that mRNA (Figure 21.10). As soon as one ribosome moves past the start codon, another ribosome can attach. A cluster of ribosomes simultaneously translating the same mRNA strand is called a polysome.
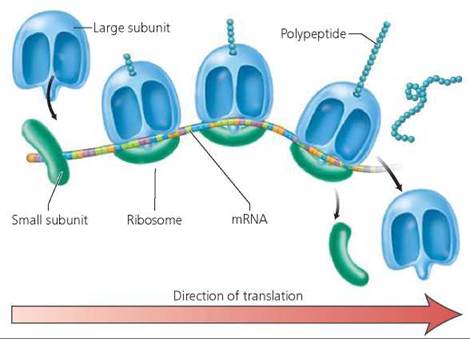
FIGURE 21.10. A polysome is a group of ribosomes reading the same mRNA molecule.
3. Termination occurs when a stop codon moves into the ribosome (Figure 21.11).
• Step 1: Stop codon moves into ribosome. There are no tRNA anticodons that pair with the stop codons, so when a stop codon moves into the ribosome, protein synthesis is terminated.
• Step 2: Parts disassemble. The newly synthesized polypeptide, the mRNA strand, and the ribosomal subunits then separate from one another.
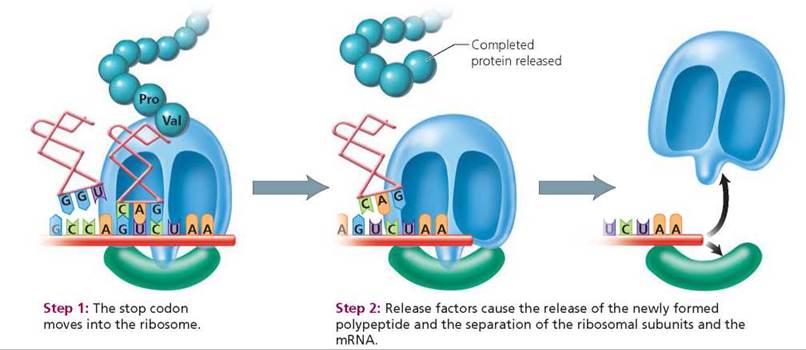
FIGURE 21.11. Termination of translation
Stop and think
Streptomycin is an antibiotic, a drug taken to slow the growth of invading bacteria and allow body defense mechanisms more time to destroy them. Streptomycin works by binding to the bacterial ribosomes and preventing an accurate reading of mRNA. Why would this process slow bacterial growth?
Mutations
DNA is remarkably stable, and the processes of replication, transcription, and translation generally occur with amazing precision. However, sometimes DNA is altered, and the alterations can change its message. Changes in DNA are called mutations. One type of mutation occurs when whole sections of chromosomes are duplicated or deleted, as discussed in Chapter 20. Now that we are familiar with the chemical structure of DNA and how it directs the synthesis of proteins, we can consider another type of mutation—a gene mutation. A gene mutation results from changes in the order of nucleotides in DNA. Although a gene mutation can occur in any cell, the only way it can be passed on to offspring is if it is present in a cell that will become an egg or a sperm. A mutation that occurs in a body cell can affect the functioning of that cell and the subsequent cells produced by that cell, sometimes with disastrous effects, but it cannot be transmitted to a person's offspring.
One type of gene mutation is the replacement of one nucleotide pair by a different nucleotide pair in the DNA double helix. During DNA replication, bases may accidentally pair incorrectly. For example, adenine might mistakenly pair with cytosine instead of thymine. Repair enzymes normally replace the incorrect base with the correct one. However, sometimes the enzymes recognize that the bases are incorrectly paired but mistakenly replace the original base (the one on the old strand) rather than the new, incorrect one. The result is a complementary base pair consisting of the wrong nucleotides (Figure 21.12).
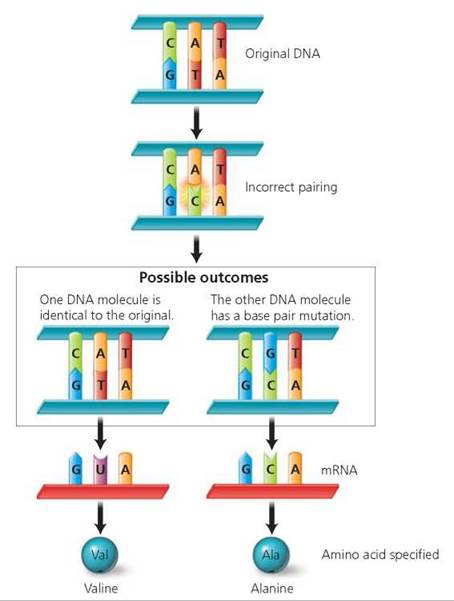
FIGURE 21.12. A base-pair substitution is a DNA mutation resulting when a base is paired incorrectly. This may change the amino acid specified by the mRNA and alter the structure of the protein.
Other types of gene mutations are caused by the insertion or deletion of one or more nucleotides. Generally, a mutation of this kind has more serious effects than does a mutation caused by substitution of one base pair for another. Recall that the mRNA is translated in units of three nucleotides (a unit called a codon). If one or two nucleotides are inserted or deleted, all the triplet codons that follow the insertion or deletion are likely to change. Consequently, mutations due to the insertion or deletion of one or two nucleotides can greatly change the resulting protein. A sentence consisting of three- letter words (representing codons) illustrates what can happen. Deleting a single letter from the sentence "The big fat dog ran" renders the sentence nonsensical:
Original: THE BIG FAT DOG RAN
After deletion of the E in THE: THB IGF ATD OGR AN
Regulating Gene Activity
At the time of your conception, you received one set of chromosomes from your father and one set from your mother. The resulting zygote then began a remarkable series of cell divisions—some of which continue in many of your body cells to this day. With each division, the genetic information was faithfully replicated, and exact copies were parceled into the daughter cells. Thus, every nucleated cell you possess, except gametes, contains a complete set of identical genetic instructions for making every structure and performing every function in your body.
How, then, can liver, bone, blood, muscle, and nerve cells look and act so differently from one another? The answer is deceptively simple: Only certain genes are active in a certain type of cell; most genes are turned off in any given cell, which leads to specialization for specific jobs. The active genes produce specific proteins that determine the structure and function of that particular cell. Indeed, as cells become specialized for specific jobs, the timing of the activity of specific genes is critical.
But what controls gene activity? The answer to this question is a bit more complex, because gene activity is controlled in several ways. Genes are regulated on several levels simultaneously.
Gene Activity at the Chromosome Level
At the chromosome level, gene activity is affected by the coiling and uncoiling of the DNA. When the DNA is tightly coiled, or condensed, the genes are not expressed. When a particular protein is needed in a cell, the region of the chromosome containing the necessary gene unwinds, allowing transcription to take place. Presumably, the uncoiling allows enzymes responsible for transcription to reach the DNA in that region of the chromosome. Other regions of the chromosome remain tightly coiled and therefore are not expressed. Indeed, the environment may affect which genes are turned on or off (see Environmental Issue essay, Environment and Epigenetics).
Regulating the Transcription of Genes
Some regions of DNA regulate the activity of other regions. As we have seen, a promoter is a specific sequence of DNA that is located adjacent to the gene it regulates. When regulatory proteins called transcription factors bind to a promoter, RNA polymerase can bind to the promoter, which begins transcription of the regulated genes.
Transcription factors can also bind to enhancers, segments of DNA that increase the rate of transcription of certain genes and, therefore, the amount of a specific protein that is produced. Enhancers also specify the timing of expression and a gene's response to external signals and developmental cues that affect gene expression.
You may recall from Chapter 10 that one of the ways certain hormones bring about their effects is by turning on specific genes. Steroid hormones, for instance, bind to receptors within a target cell. The hormone-receptor complex then finds its way to the chromatin in the nucleus and turns on specific genes. For example, one such complex turns on the genes in cells that produce facial hair—explaining why your father may have a beard but your mother probably does not, even though she has the necessary genes to grow one. In this case, the sex hormone testosterone binds to a receptor and turns on hair-producing genes. Facial hair follicle cells of both men and women have the necessary testosterone receptors. However, women usually do not produce enough testosterone to activate the hair-producing genes, so bearded women are rare.
Stop and think
Why do female athletes who inject themselves with testosterone to stimulate muscle development sometimes develop increased facial hair?
Environmental Issue
Environment and Epigenetics
Your lifestyle may influence the health of your great grandchild. How is this possible? It can occur through epigenetics, which involves a stable alteration in gene expression without changes in DNA sequence. In other words, it regulates how genes are expressed without changing the proteins they encode. We will consider two epigenetic processes: DNA methylation and histone acetylation. These processes alter gene expression by affecting how tightly packaged the DNA molecule is. DNA is packaged with proteins to form chromosomes. DNA methylation (adding a methyl group to the cytosine bases in DNA) turns off the activity of a gene by bringing in proteins that act to compact DNA into a tighter form. On the other hand, histone acetylation makes the DNA less tightly coiled and gene expression easier.
We now know that these processes can be affected by the environment and that the pattern of DNA methylation is dynamic and changes over time. DNA methylation patterns can be affected by environmental factors, cause disease, be transmitted through generations, and, potentially, influence evolution. DNA is sensitive to the environment, so what we eat and the chemicals we are exposed to, including pesticides, tobacco smoke, hormones, and nutrients, may influence our health by affecting our gene expression patterns. For example, maternal nutrition during pregnancy can cause epigenetic changes in gene activity in the fetus that may increase susceptibility to obesity, type-2 diabetes, heart disease, and cancer. The quantity of food consumed during pregnancy alters the offspring's susceptibility to cardiovascular disease. Epigenetics is also thought to play a role in human behavioral disorders, such as autism spectrum disorders (discussed in Chapter 18a), Rett syndrome (a developmental disorder that affects the nervous system), and Fragile-X syndrome (an inherited form of mental impairment). For example, there is some evidence that a gene needed to respond to oxytocin (a hormone important in social bonding) is turned off in some people with autism. As we will see in Chapter 21a, cancer development is controlled by cancer-inhibiting and cancer-promoting genes. If cancer-inhibiting genes are turned off or cancer-promoting genes turned on cancer can result. Changes in the pattern of gene expression are found in cancers of the cervix, prostate, breast, stomach, and colon.
Although DNA methylation patterns are considered to be stable, some studies suggest that methylation can be reversed in adulthood. Foods such as broccoli, onions, and garlic may reduce methylation, allowing genes to be expressed. Researchers are actively looking for drugs that will alter the pattern of methylation and cure cancer.
Questions to Consider
• Do you think that epigenetics increases or decreases a person's responsibility for their own behavior?
• Researchers may someday develop an “epigenetic diet” that favors positive changes in gene activity. Would you follow that diet? Do you think that pregnant women should be required to follow that diet?
Genetic Engineering
The manipulation of genetic material for human purposes, a practice called genetic engineering, began almost as soon as scientists started to understand the language of DNA. Genetic engineering is part of the broader endeavor of biotechnology, a field in which scientists make controlled use of living cells to perform specific tasks. Genetic engineering has been used to produce pharmaceuticals and hormones, improve diagnosis and treatment of human diseases, increase food production from plants and animals, and gain insight into the growth processes of cells.
Recombinant DNA
The basic idea behind genetic engineering is to put a gene of interest—in other words, one that produces a useful protein or trait—into another piece of DNA to create recombinant DNA, which is DNA combined from two or more sources. The recombinant DNA, carrying the gene of interest, is then placed into a rapidly multiplying cell that quickly produces many copies of the gene. The final harvest may consist of large amounts of the gene product or many copies of the gene itself. Let's take a closer look at the procedure one step at a time.
1. The gene of interest is sliced out of its original organism and spliced into vector DNA. Both the DNA originally containing the gene of interest and the vector DNA, which receives the transferred genes and transports it to a new cell, are cut at specific sequences that are recognized by a restriction enzyme. This is a type of enzyme that makes a staggered cut between specific base pairs in DNA, leaving several unpaired bases on each side of the cut. There are many kinds of restriction enzymes; each kind recognizes and cuts a different sequence of DNA. The stretch of unpaired bases produced on each side of the cut is called a sticky end because of its tendency to pair with the single-stranded stretches of complementary base sequences on the ends of other DNA molecules that were cut with the same restriction enzyme (Figure 21.13).
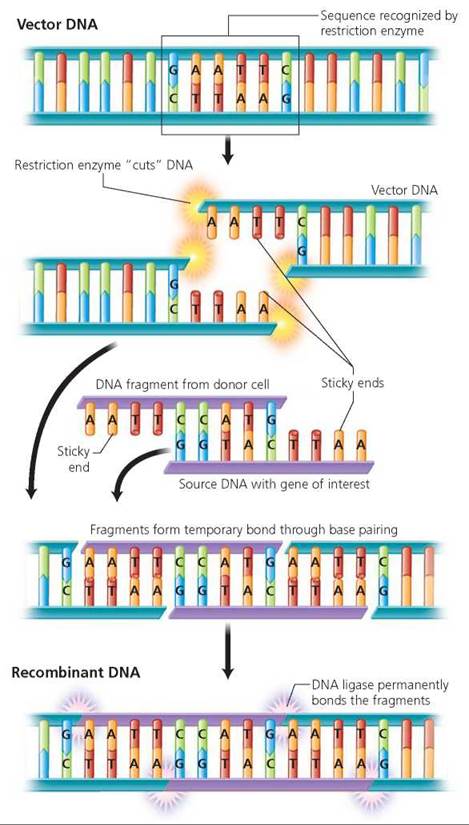
FIGURE 21.13. DNA from different sources can be spliced together using a restriction enzyme to make cuts in the DNA. A restriction enzyme makes a staggered cut at a specific sequence of DNA, leaving a region of unpaired bases on each cut end. The region of singlestranded DNA at the cut end is called a sticky end, because it tends to pair with the complementary sticky end of any other piece of DNA that has been cut with the same restriction enzyme, even if the pieces of DNA came from different sources.
The sticky ends are the secret to splicing the gene of interest and the vector DNA. The sticky ends of DNA from different sources will be complementary and stick together as long as they have been cut with the same restriction enzyme. The initial attachment between sticky ends is temporary, but the ends can be "pasted" together permanently by another enzyme, DNA ligase. The resulting recombinant DNA contains DNA from two sources.
2. The vector is used to transfer the gene of interest to a new host cell. Biological carriers that ferry the recombinant DNA to a host cell are called vectors. A common vector is bacterial plasmids, which are small, circular pieces of self-replicating DNA that exist separately from the bacterial chromosome.2 As previously described, the source DNA (that is, the source of the gene of interest) and the plasmid (vector) DNA are both treated with the same restriction enzyme. Afterward, fragments of source DNA, some of which will contain the gene of interest, will be incorporated into plasmids when their sticky ends join. The recombinant DNA is mixed with bacteria in a test tube. Under the right conditions, some of the bacterial cells will then take up the recombined plasmids (Figure 21.14).
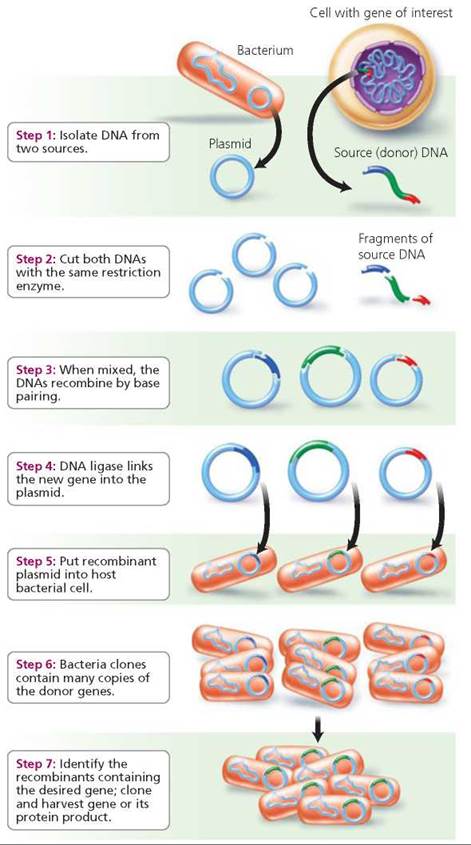
FIGURE 21.14. An overview of genetic engineering using plasmids
Although the basic strategy is usually the same, there are many variations on this theme of transporting a gene into a new host. For instance, the gene of interest is sometimes combined with viral DNA. The viruses are then used as vectors to insert the recombinant DNA into a host cell. Cells other than bacteria, including yeast or animal cells, can also be used as vectors.
3. The recombinant organism containing the gene of interest is identified and isolated from the mixture of recombinants. When plasmids are used as vector DNA, each recombinant plasmid is introduced into a single bacterial cell, and each cell is then grown into a colony. Each colony contains a different recombinant plasmid. The bacteria containing the gene of interest must be identified and isolated.
4. The gene is amplified through bacterial cloning or by use of a polymerase chain reaction. After the colony containing the gene of interest has been identified, researchers usually amplify (that is, replicate) the gene, producing numerous copies. Gene amplification is accomplished using one of two techniques: bacterial cloning or a polymerase chain reaction.
Cloning. Bacteria containing the plasmid with the gene of interest can be grown in huge numbers by cloning. Each bacterium divides many times to form a colony. Thus, each colony constitutes a clone—a group of genetically identical organisms all descended from a single cell. In this case, all the members of the clone carry the same recombinant DNA. Later, the plasmids can be separated from the bacteria, a process that partially purifies the gene of interest. The plasmids then can be taken up by other bacteria that will thus become capable of performing a service deemed useful by humans. Alternatively, the plasmids can be transferred into plants or animal cells— creating transgenic organisms—organisms containing genes from another species.
Polymerase chain reaction (PCR). In PSR (Figure 21.15), the DNA of interest is unzipped, by gentle heating, to break the hydrogen bonds and form single strands. The single strands, which will serve as templates, are then mixed with primers— special short pieces of nucleic acid—one primer with bases complementary to each strand. The primers serve as start tags for DNA replication. Nucleotides and a special heat-resistant DNA polymerase, which promotes DNA replication, are also added to the mixture, which is then cooled to allow base pairing. Through base pairing, a complementary strand forms for each single strand. The procedure is then repeated many times, and each time the number of copies of the DNA of interest is doubled. In this way, billions of copies of the DNA of interest can be produced in a short time.
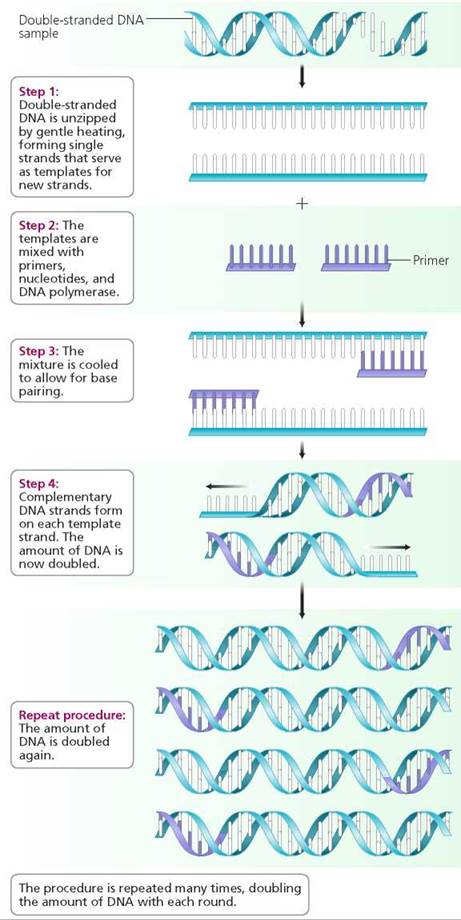
FIGURE 21.15. The polymerase chain reaction (PCR) rapidly produces a multitude of copies of a single gene or of any desired segment of DNA. PCR amplifies DNA more quickly than does bacterial cloning. It has many uses besides genetic engineering, including DNA fingerprinting.
What would you do?
Genetic engineering involves altering an organism's genes—adding new genes and traits to microbes, plants, or even animals. Do you think we have the right to “play God” and alter life-forms in this way? The U.S. Supreme Court has approved patenting of genetically engineered organisms, first of microbes and now of mammals such as pigs that are genetically modified for use in organ transplant. If you were asked to decide whether it is ethical to patent a new life-form, how would you respond?
Applications of Genetic Engineering
Genetic engineering has been used in two general ways.
• Genetic engineering provides a way to produce large quantities of a particular gene product. The useful gene is transferred to another cell, usually a bacterium or a yeast cell, that can be grown easily in large quantities. The cells are cultured under conditions that cause them to express the gene, after which the gene product is harvested. For example, genetically engineered bacteria have been used to produce large quantities of human growth hormone (Figure 21.16). Treatment with growth hormone allows children with an underactive pituitary gland to grow to nearly normal height.
• Genetic engineering allows a gene for a trait considered useful by humans to be taken from one species and transferred to another species. The transgenic organism then exhibits the desired trait. For example, scientists have endowed salmon with a gene from an eel-like fish. This gene causes the salmon to produce growth hormone year- round (something they do not normally do). As a result, the salmon grow faster than normal.
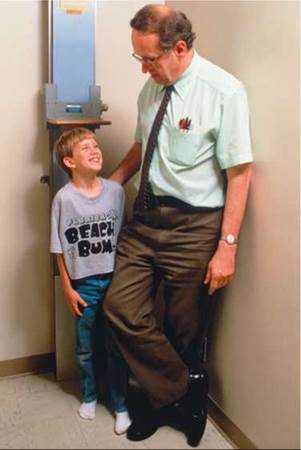
FIGURE 21.16. Genetic engineering is used to produce large quantities of a desired protein or to create an organism with a desired trait. This boy has an underactive pituitary gland. Its undersecretion of growth hormone would have caused him to be very short, even as an adult. However, growth hormone from genetically engineered bacteria has helped him grow to an almost normal height.
Environmental applications. Genetic engineering also has environmental applications. For example, in sewage treatment plants, genetically engineered microbes lessen the amount of phosphate and nitrate discharged into waterways. Phosphate and nitrate can cause excessive growth of aquatic plants, which could choke waterways and dams, and of algae, which can produce chemicals that are poisonous to fish and livestock. Microorganisms are also being genetically engineered to modify or destroy chemical wastes or contaminants so that they are no longer harmful to the environment. For instance, oil-eating microbes that can withstand the high salt concentrations and low temperatures of the oceans have proven useful in cleaning up after marine oil spills.
Livestock. Genetic engineering has also been used on livestock. Genetically engineered vaccines have been created to protect piglets against a form of dysentery called scours, sheep against foot rot and measles, and chickens against bursal disease (a viral disease that is often fatal). Genetically engineered bacteria produce bovine somatotropin (BST), a hormone naturally produced by a cow's pituitary gland that enhances milk production. Injections of BST can boost milk production by nearly 25%.
Transgenic animals have been created by injecting a fertilized egg with the gene of interest in a petri dish. The goals of creating transgenic animals include making animals with leaner meat, sheep with softer wool, cows that produce more milk, and animals that mature more quickly.
Pharmaceuticals. Genes have been put into a variety of cells, ranging from microbes to mammals, to produce proteins for treating allergies, cancer, heart attacks, blood disorders, autoimmune disease, and infections.
Genetically engineered bacteria have also been used to create vaccines for humans. You may recall from Chapter 13 that a vaccine typically uses an inactivated bacterium or virus to stimulate the body's immune response to the active form of the organism. The idea is that the body will learn to recognize proteins on the surface of the infectious organism and mount defenses against any organism bearing those proteins. Because the organism used in the vaccine was rendered harmless, the vaccine cannot trigger an infection. Scientists produce genetically engineered vaccines by putting the gene that codes for the surface protein of the infectious organism into bacteria. The bacteria then produce large quantities of that protein, which can be purified and used as a vaccine. The vaccine cannot cause infection, because only the surface protein is used instead of the infectious organism itself.
Plants have also been used to produce therapeutic proteins. Engineered bananas that produce an altered form of the hepatitis B virus surface protein are being developed as an edible vaccine against the liver disease hepatitis B. Someday, you may eat a banana in order to be vaccinated, instead of receiving an injection against hepatitis B. Plants are also being engineered to produce "plantibodies," antibodies made by plants. For example, soybeans are being cultivated that contain human antibodies to the herpes simplex virus that causes genital herpes. A human gene for an antibody that binds to tumor cells has been transplanted into corn. The antibodies can then deliver radioisotopes to cancer cells, selectively killing them.
Pharming is a word that comes from the combination of the words farming and pharmaceuticals. In gene pharming, transgenic animals are created that produce a protein with medicinal value in their milk, eggs, or blood. The protein is then collected and purified for use as a pharmaceutical. When the pharm animal is a mammal, the gene is expressed in mammary glands. The desired protein is then extracted and purified from milk (Figure 21.17). For example, the gene for the protein alpha-1-antitrypsin (AAT) has been inserted into sheep that then secrete AAT in their milk. People with an inherited, potentially fatal form of emphysema (a lung disease) take AAT as a drug. It is also being tested as a drug to prevent lung damage in people with cystic fibrosis. The first drug made from the milk of a transgenetic goat was an anticlotting drug called ATryn. It is given to people with a blood-clotting deficiency when they must undergo surgery. Researchers have also created a transgenic goat to produce milk containing lysozyme, an antibacterial agent. Lysozyme can be used to treat intestinal infections that kill millions of children in underdeveloped countries.
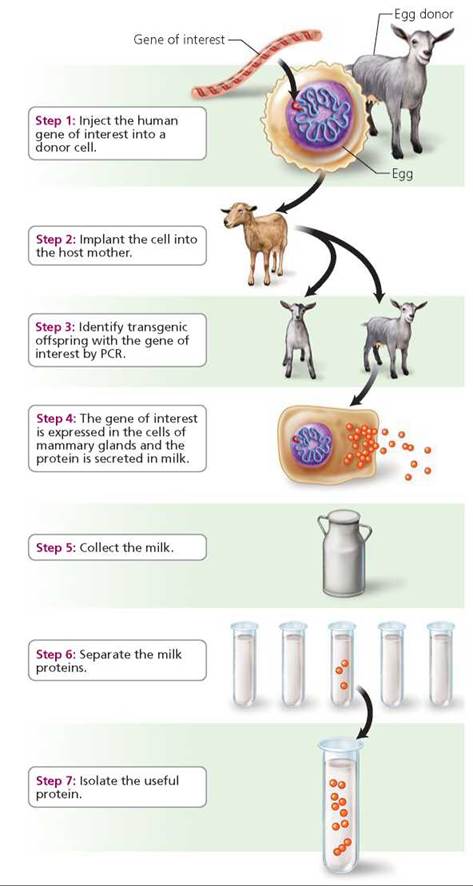
FIGURE 21.17. The procedure for creating a transgenic animal that will produce a useful protein in its milk
Raw materials. Genetic engineering is also used to produce useful new raw materials. For example, spider silk is five times stronger than steel and still lightweight. Attempts have been made to farm spiders for their silk, but spiders are too aggressive to live close together. Goats have been genetically engineered to possess the gene for spider silk and secrete spider silk proteins in their milk (Figure 21.18). More recently, E. coli bacteria have been genetical engineered to produce spider silk proteins. The spider silk proteins can be spun into a fine thread, which could be used for bulletproof clothing, thinner surgical thread, and stronger yet lighter racing cars and aircraft.

FIGURE 21.18. This transgenic goat has the gene for making spider silk, one of the strongest substances known. The spider silk protein can be extracted from the goat’s milk and spun into threads that can be used for products in which strength and light weight are important qualities.
Agriculture. Most of us experience some of the results of genetic engineering at our dinner tables (see Health Issue Essay, Genetically Modified Food.) The most common traits that have been genetically engineered into crops are resistance to pests and resistance to herbicides. Scientists also have developed two virus-resistant strains of papaya and distributed them to papaya growers in Hawaii, saving the industry from ruin. In addition, different strains of rice have been genetically engineered to resist disease-causing bacteria and to withstand flooding of the paddy. Other plants have been genetically engineered to be more nutritious. For example, golden rice is a strain of rice that has been genetically engineered to produce high levels of beta-carotene, a precursor of vitamin A, which is in short supply in certain parts of the world. More than 100 million children worldwide suffer from vitamin A deficiency, and 500,000 of them go blind every year because of that deficiency. Although golden rice cannot supply a complete recommended daily dose of vitamin A, the amount it contains could be helpful to a person whose diet is very low in vitamin A. Other crops have also been created that grow faster, produce greater yields, and have longer shelf lives.
Gene Therapy
The problems associated with many genetic diseases arise because a mutant gene fails to produce a normal protein product. The goal of gene therapy is to cure genetic diseases by putting normal, functional genes into the body cells that were affected by the mutant gene. The functional gene would then produce the needed protein.
Methods of delivering a healthy gene. One way a healthy gene can be transferred to a target cell is by means of viruses. Viruses generally attack only one type of cell. For instance, an adenovirus, which causes the common cold, typically attacks cells of the respiratory system. A virus consists largely of genetic material, commonly DNA, surrounded by a protein coat (see Chapter 13a). Once inside a cell, the viral DNA uses the cell's metabolic machinery to produce viral proteins. If a healthy gene is spliced into the DNA of a virus that has first been rendered harmless, the virus will deliver the healthy gene to the host cell and cause the desired gene product to be produced (Figure 21.19).
Another type of virus used in gene therapy is a retrovirus, a virus whose genetic information is stored as RNA rather than DNA. Once inside the target cell, a retrovirus rewrites its genetic information as double-stranded DNA and inserts the viral DNA into a chromosome of the target cell.
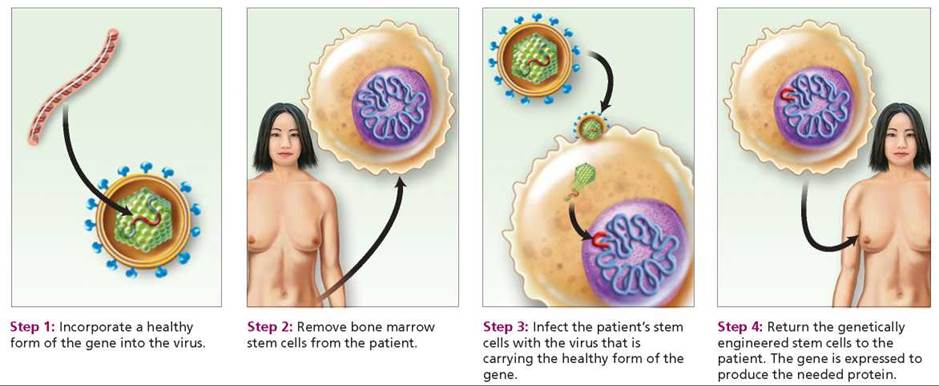
FIGURE 21.19. Gene therapy using a virus. In gene therapy, a healthy gene is introduced into a patient who has a genetic disease caused by a faulty gene.
Environmental Issue
Genetically Modified Food
From dinner tables to diplomatic circles, people are discussing genetically modified (GM) food. This relatively recent interest is somewhat ironic, considering that people in the United States have been eating GM food since the mid-1990s. More than 70% of processed foods sold in the United States contain genetically modified ingredients. Yet many people vehemently object to GM food.
Why is something as common as GM food controversial? The concerns about it can generally be divided into three categories: health issues, social issues, and environmental issues. Let's explore these categories one at a time.

Health Concerns
A panel of the National Academy of Sciences (NAS) has issued a report saying that genetically engineered crops do not pose health risks that cannot also be caused by crops created by conventional breeding. However, because genetic engineering could produce unintended harmful changes in food, the NAS panel recommends scrutiny of GM foods before they can be marketed. Currently, the U.S. Department of Agriculture, the Food and Drug Administration (FDA), and the Environmental Protection Agency regulate genetically modified foods. The NAS panel concluded that the GM foods already on the market are safe.
· The larger salmon in the back of the photo has been genetically modified to grow faster than the normal salmon in front.
A common safety concern is that GM foods may contain allergens (substances that cause allergies). After a protein is produced, the cell modifies it in various ways. The protein may be modified in the genetically modified plant differently from the way it would in an unmodified cell, and the modification could produce an allergen. The likelihood that this may occur is reduced by rigorous testing. Most known allergens share certain properties. They are proteins, relatively small molecules, and resistant to heat, acid, and digestion in the stomach. If a protein produced by a GM plant has any of the properties typical of an allergen or is structurally similar to a known allergen, the FDA considers it to be a potential allergen and requires that the protein undergo additional allergy testing.
Bacterial resistance to antibiotics is likely to be a major threat to public health in this decade (see Chapter 13a). When bacteria are resistant to an antibiotic, the drug will not kill them and thus will no longer cure the human disease for which they are the cause. Some people worry about the scientific practice of putting genes for resistance to an antibiotic into GM crops as markers to identify the plants with the modified genes. Plant seedlings thought to be genetically modified are grown in the laboratory in the presence of an antibiotic. Only those seedlings with the gene for resistance will survive. Because of the way they were engineered, the surviving plants also contain the “useful” gene.
What worries some people is that the genes for resistance to antibiotics could be transferred to bacteria, making the bacteria resistant to antibiotics. The receiving bacteria might be those that normally live in the human digestive system, or they might be bacteria ingested with food. It is not known whether genes can be transferred from a plant to a bacterium. However, it is known that bacteria can easily and quickly transfer genes for antibiotic resistance to one another. Thus, a harmless bacterium in the gut could transfer the gene for antibiotic resistance to a disease-causing bacterium.
The transfer of antibiotic-resistance genes from GM plants to bacteria could have serious consequences. For this reason, antibiotic- resistance marker genes are being phased out in favor of other marker genes, such as a green fluorescent protein. Scientists also have developed a way to inactivate the antibiotic-resistance gene if it were to be transferred to bacteria.
Environmental Concerns
Proponents of GM foods argue that herbicide- resistant and pesticide-resistant crops reduce the need for spraying with herbicides and pesticides. So far, experience has shown that the validity of this argument depends on the crop. Pest-resistant cotton has substantially reduced the use of pesticides, but pest-resistant corn probably has not. Farmers who grow herbicide- resistant crops still spray with herbicides, but they change the type of herbicide they use to a type that is less harmful to animals.
Unfortunately, engineered crops containing insecticides could have undesirable effects on insects. Genetically engineering insecticides into plants could hasten the development of insect resistance to that insecticide, making the insecticide ineffective—not just for the genetically modified crop, but for all crops.
Another concern is that genetically modified organisms could harm other organisms. One example is that pollen from pest-resistant corn has been shown to harm monarch butterfly caterpillars. Fortunately, monarch caterpillars rarely encounter enough pollen to be harmed, and most of the pest-resistant corn grown in the United States today does not produce pollen that is harmful to monarch butterflies. A second example of a genetically modified organism that has the potential to harm other organisms is the salmon that produce more growth hormone and grow several times faster than their wild relatives do. These salmon are grown on fish farms. If the FDA grants approval, the gene-altered salmon could dramatically cut costs for fish farmers and consumers. However, when the genetically modified salmon are grown in tanks with ordinary salmon and food is scarce, the genetically modified salmon eat most of the food and some of their ordinary companions. What would happen if the genetically modified salmon escaped from their pens on the fish farm? They might mate with the wild salmon and create less healthy offspring or outcompete wild salmon for food, which could eventually cause the extinction of the wild salmon. Escape is possible. During the past few years, hundreds of thousands of fish have escaped from fish farms when floating pens were ripped apart by storms or sea lions. To minimize the risk that genetically modified salmon could destroy the population of wild salmon, scientists plan to breed the fish inland, sterilize the offspring, and ship only sterile fish to coastal pens. The sterilization procedure is effective in small batches of fish, but it is not known whether the procedure is completely effective in large batches of fish.
Critics of GM foods also fear that crops genetically engineered to resist herbicides could become "super weeds" that could not be controlled with existing chemicals. In North Dakota, GM canola plants that are resistant to herbicides are growing along roadsides. Canola can hybridize with at least two wild weeds. The two original strains of GM canola were each resistant to a different herbicide. As a result of cross-pollination, some of the canola plants found in the wild are resistant to both herbicides, which suggests that the GM traits are stable in the wild and are evolving.
Social Concerns
Proponents of GM food claim that GM food can assist in the battle against world hunger. We have seen that genetic engineering can produce crops that resist pests and disease. It can also produce crops with greater yields and crops that will grow in spite of drought, depleted soil, or excess salt, aluminum, or iron. Foods can also be genetically modified to contain higher amounts of specific nutrients.
Critics of using GM food to battle world hunger argue that the problem of hunger has nothing to do with an inability to produce enough food. The problem, they say, is a social one of distributing food so that it is available to the people who need it.
Some developing countries are resisting the use of GM seeds. Part of the resistance stems from lingering health concerns. However, many farmers in developing countries also object to GM seeds because the GM plants do not seed themselves. The need to buy seeds each year places a financial burden on poor farmers.
Questions to Consider
• Do you consider GM food to be a blessing or a danger to the world? What are your reasons?
• If a genetically modified organism that was created for food begins to cause environmental problems, who should be held accountable?
• Do you think that foods containing genetically modified components should be labeled as such?
Gene therapy results. More than 4000 human diseases have been traced to defects in single genes. Although the Food and Drug Administration has not yet approved a gene therapy for any of these conditions, hundreds of clinical trials of gene therapies are currently under way, including trials of possible therapies for cystic fibrosis and cancer (discussed in Chapter 21a).
The first condition to be treated experimentally with gene therapy was a disorder referred to as severe combined immunodeficiency disease (SCID). The immune system of children with SCID is nonfunctional, leaving them vulnerable to infections. The cause of the problem is a mutant gene that prevents the production of an enzyme called adenosine deaminase (ADA). Without ADA, white blood cells never mature—they die while still developing in the bone marrow. The first gene therapy trial began in 1990, when white blood cells of a 4-year-old SCID patient, Ashanthi DeSilva, were genetically engineered to carry the ADA gene and then returned to her tiny body. Her own gene-altered white blood cells began producing ADA, and her body defense mechanisms were strengthened. Ashanthi's life began to change. She was not ill as often as she had been before. She could play with other children. However, the life span of white blood cells is measured in weeks, and when the number of gene-altered cells declined, new gene-altered cells had to be infused. Ashanthi is now in her twenties and has a reasonably healthy immune system. However, she still needs repeated treatments.
French scientists believe they have cured 10 children with X-SCID by using gene therapy (Figure 21.20). X-SCID is a severe combined immunodeficiency syndrome caused by a mutant gene on the X chromosome. It is still too soon to know for certain whether all 10 children will require treatment in the future. However, four children in the French studies who had shown improvement in symptoms of X-SCID developed leukemia from the therapy.

FIGURE 21.20. Rhys Evans is the first person to be cured of X-linked severe combined immunodeficiency disease (X-SCID) by gene therapy. The immune system of a person with X-SCID is nonfunctional. Rhys Evans’s immune system was strengthened, and he can now go to public places without fearing contact with people who might carry germs. He can play with other children.
A form of muscular dystrophy has recently been treated with gene therapy. Researchers packed a virus with the normal form of the protein that is deficient in this form of muscular dystrophy. The virus was then injected directly into the muscles. The level of the protein and the gene expression of the protein remained high for several months, restoring some muscle function to the patients.
What would you do?
Without treatment, children with X-SCID die at a young age. The gene therapy treatment for X-SCID that caused leukemia in four French boys does appear to have cured this deadly disorder in other patients. If you had a child with X-SCID, would you want him to have this gene therapy treatment? Why or why not? What factors would you consider in making your decision?
Genomics
A genome is the entire set of genes carried by one member of a species—in our case, one person. Genomics is the study of entire genomes and the interactions of the genes with one another and the environment.
Human Genome Project
One goal of genomics is to determine the location and sequences of genes. Researchers have developed supercomputers that automatically sequence (determine the order of bases in) DNA. The supercomputers were put to use on a massive scale in the Human Genome Project, a worldwide research effort, completed in 2003, to sequence the human genome. As a result, we now have some idea of the locations of genes along all 23 pairs of human chromosomes and the sequence of the estimated 3 billion base pairs that make up those chromosomes. Although the exact number of human genes is still not known for certain, scientists now estimate that the human genome consists of 20,000 to 25,000 genes, not 100,000 as originally thought. One reason for the smaller number of actual genes is that many gene families have related or redundant functions and therefore are able to share certain genes, so that fewer are needed to carry out all the body's functions. A second reason is that many genes are now known to code for parts of more than one protein.
In addition, researchers have identified and mapped to specific locations on specific chromosomes genes for more than 1400 genetic diseases. It is hoped that this information will give scientists a greater ability to diagnose, analyze, and eventually treat many of the 4000 diseases of humans that have a known genetic basis. Researchers have already cloned the genes responsible for many genetic diseases, including Duchenne muscular dystrophy, retinoblastoma, cystic fibrosis, and neurofibromatosis. These isolated genes can now be used to test for the presence of the same disease-causing genes in specific individuals. As we saw in Chapter 20, some gene tests can be used to identify people who are carriers for certain genetic diseases such as cystic fibrosis, allowing families to make choices based on known probabilities of bearing an affected child; similar tests can be used for prenatal diagnosis and for diagnosis before symptoms of the disease begin. After a disease-related gene has been identified, scientists can study it to learn more about the protein it codes for and perhaps discover ways to correct the problem.
We have also learned from the Human Genome Project that humans are identical in 99.9% of the sequences of their genes. As scientists gain greater understanding of the 0.1% of DNA that differs from person to person, they expect to learn more about why some people develop heart disease, cancer, or Alzheimer's disease and others do not.
Microarray Analysis
A second goal of genomics is to understand the mechanisms that control gene expression. More than 95% of human DNA does not code for protein; however, some of these noncoding DNA sequences function as regulatory regions that determine when, where, and how much of certain proteins are produced. Because gene activity plays a role in many diseases, the study of how these regions turn genes on or off may lead to advances in diagnosis and treatment.
One of the tools researchers use in this effort is the microarray, which consists of thousands of DNA sequences stamped onto a single glass slide called a DNA chip. Researchers use microarrays to monitor large numbers of DNA segments to discover which genes are active and which are turned off under different conditions, such as in different tissue types, different stages of development, or in health and disease. For example, they may use microarrays to identify genes that are active in cancerous cells but not in healthy cells (Figure 21.21). Presumably, the genes that are active in cancerous cells play a role in the development of cancer.
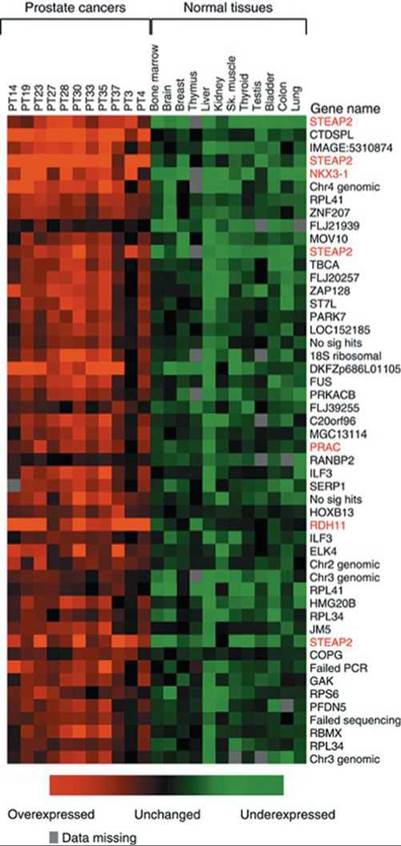
FIGURE 21.21. A comparison of microarrays showing the pattern of gene activity in prostate cancer and in normal tissue. The genes that are active in tissue with prostate cancer but not in normal tissue probably play a role in the development of cancer.
Besides identifying gene activity in health and disease, microarray analysis is useful in identifying genetic variation in the members of a population. Some of these genetic differences are in the form of single-nucleotide polymorphisms (SNPs, or snips). These are DNA sequences that can vary by one nucleotide from person to person, and the differences in their protein products are thought to influence how we respond to stress and diseases, among other things. As researchers learn more about SNPs, they may be able to develop treatments tailored to the genetic makeup of each individual. These are the kinds of discoveries that can open the door for gene therapy. While individualized gene therapy may be useful in the future, we already use identification of individual differences in DNA on a regular basis in DNA fingerprinting (see the Ethical Issue essay, Forensic Science, DNA, and Personal Privacy).
What would you do?
Some people worry that once we know the location and function of every gene and have perfected the techniques of gene therapy, we will no longer limit gene manipulation to repairing faulty genes but will begin to modify genes to enhance human abilities. Should people be permitted to design their babies by choosing genes that they consider superior? What do you think? Where should the line be drawn? Who should draw that line? Who should decide which genes are “good?”
When SNPs are located near one another on a chromosome, they tend to be inherited together. A group of SNPs in a region of a chromosome is called a haplotype. The International HapMap Project is a scientific consortium whose purpose is to describe genetic variation between populations. Researchers collaborating on this project compare haplotype frequencies in groups of people who have a certain disease to those of a group without the disease, hoping to identify genes associated with the disease.
Comparison of Genomes of Different Species
The DNA of certain widely studied organisms, including the mouse, the fruit fly, a roundworm, yeast, slime mold, and the honeybee have also been mapped. From these genomes, geneticists hope to gain some insight into basic biology, including basic principles of the organization of genes within the genome, gene regulation, and molecular evolution. Humans share many genes with other organisms. For example, we share 50% of our genes with the fruit fly and 90% of our genes with the mouse. These genetic similarities are evidence of our common evolutionary past. The genes and genetic mechanisms we share with other organisms are likely to be important in determining body form as well as influencing development and aging.
Looking ahead
In Chapter 19, we learned about the cell cycle. In Chapters 20 and 21, we learned about genes, their inheritance, and their regulation, and we also considered how mutations affect gene functions. In Chapter 21a, we use this information to understand cancer, a family of diseases in which mutations in genes that regulate the cell cycle cause a loss of control over cell division.
Ethical Issue
Forensic Science, DNA, and Personal Privacy
So-called DNA fingerprints, like the more conventional prints left by fingers, can help identify the individuals they belong to out of a large population. DNA fingerprinting refers to techniques of identifying individuals on the basis of unique features of their DNA. DNA fingerprints are possible because many regions of DNA are composed of small, specific sequences of DNA that are repeated many times. Most commonly used are repeated units of 1 to 5 bases, which are called short tandem repeats (STRs). The number of times these sequences are repeated varies considerably from person to person, from a few to 100 repeats. Because of these differences, the segments can be used to match a sample of DNA to the person whose cells produced the sample.
The first step in preparing a DNA fingerprint is to extract DNA from a tissue sample. The type of tissue does not matter, and one type of tissue can be successfully compared to another type. Commonly used sources include blood, semen, skin, and hair follicles, because they are not too painful to remove, they are readily available, or they are left at a crime scene.
First, the amount of DNA is greatly increased using PCR, as described in Figure 21.15. The primers used are sequence specific for the regions on either side of the repeating region. This produces many copies of the repeating region, which are then analyzed to determine the number of repeats present.
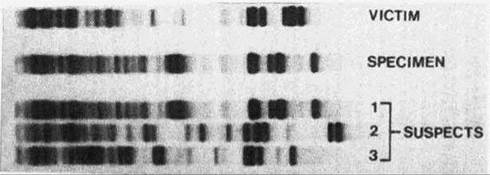
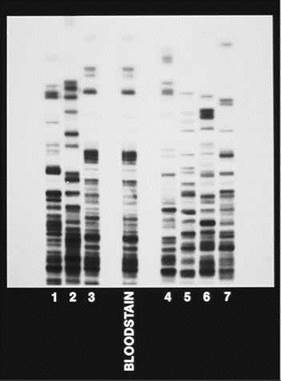
FIGURE 21.A. The pattern of banding in a DNA fingerprint is determined by the sequence of bases in a person’s DNA and is, therefore, unique to each person. A match between DNA fingerprints can identify the source of a tissue sample from a crime scene with a high degree of certainty. Which suspect’s DNA fingerprint matches this specimen found at a crime scene?
The FBI uses 13 STRs as a core set for forensic analysis. The resulting DNA fingerprint is unique to the person who produced the DNA. Moreover, any DNA sample taken from the same person would always be identical. But the fingerprint profiles resulting from the DNA of different people are always different (except perhaps for identical siblings), because the number and sizes of the fragments are determined by the unique sequence of bases in each person’s DNA.
DNA fingerprinting has many applications, but the most familiar is probably its use in crime investigations. In these cases the DNA fingerprint is usually created from a sample of tissue, such as blood or hair follicles, collected at the crime scene. A fingerprint can be produced from tissue left at the scene years before. This fingerprint is then compared with the DNA fingerprints of various suspects. A match reveals, with a high degree of certainty, the person who was the source of the sample from the crime scene (Figure 21.A).
Of course, the degree of certainty of a match between DNA fingerprints depends on how carefully the analysis was done. Because DNA fingerprints are being used as evidence in an increasing number of court cases each year, it is important that national standards be set to ensure the reliability of these molecular witnesses. It is generally easier to declare with certainty that two DNA fingerprints do not match than it is to be sure that they do. In the United States more than 200 convicts have been found innocent through DNA testing during the last decade.
Has DNA testing gone too far? All states in the United States collect DNA samples from people convicted of sex crimes and murder. Several other states also collect DNA samples from people convicted of other felonies, such as robbery. About 30 states collect DNA samples from people accused of misdemeanors, including loitering, shoplifting, or vandalism. In many cases, the DNA is stored in a database, even if the person is found innocent of the crime. People who are simply cooperating with the investigation may also provide DNA samples. These, too, are added to a national database.
Questions to Consider
• Do you think everyone arrested of a crime should have the right to DNA fingerprinting to prove his or her innocence? If so, who should pay for the process?
• Is the creation of a national database of DNA fingerprints an invasion of privacy? Is it any more so than a database of actual fingerprints or mug shots?
• Under what conditions do you think DNA samples should be obtained?
Highlighting the Concepts
Form of DNA (pp.434-435)
• DNA consists of two strands of nucleotides linked by hydrogen bonds and twisted together to form a double helix. Each nucleotide consists of a phosphate, a sugar called deoxyribose, and one of four nitrogenous bases: adenine, thymine, cytosine, or guanine. The sugar and phosphate components of the nucleotides alternate along the two sides of the molecule. Pairs of bases meet and form hydrogen bonds in the interior of the double helix; these pairs resemble the rungs on a ladder.
• According to the rules of complementary base pairing, adenine binds only with thymine, and cytosine binds only with guanine.
Replication of DNA (pp. 435-436)
• DNA replication is semiconservative. Each new double-stranded DNA molecule consists of one old and one new strand. The enzyme DNA polymerase "unzips" the two strands of a molecule (the parent molecule), allowing each strand to serve as a template for the formation of a new strand. Complementary base pairing ensures the accuracy of replication.
Gene Expression (pp. 436-441)
• Genetic information is transcribed from DNA to RNA and then translated into a protein.
• Transcription is the synthesis of RNA by means of base pairing on a DNA template. RNA differs from DNA in that the sugar ribose replaces deoxyribose, and the base uracil replaces thymine. Most RNA is single stranded.
• Messenger RNA (mRNA) carries the DNA genetic message to the cytoplasm, where it is translated into protein. The genetic code is read in sequences of three RNA nucleotides; each triplet is called a codon. Each of the 64 codons specifies a particular amino acid or indicates the point where translation should start or stop.
• Transfer RNA (tRNA) interprets the genetic code. At one end of the tRNA molecule is a sequence of three nucleotides called the anticodon that pairs with a codon on mRNA in accordance with basepairing rules. The tail of the tRNA binds to a specific amino acid.
• Each of the two subunits of a ribosome consists of ribosomal RNA (rRNA) and protein. A ribosome brings tRNA and mRNA together for protein synthesis.
• Translation of the genetic code into protein begins when the two ribosomal subunits and an mRNA assemble, with the mRNA sitting in a groove between the ribosome's two subunits. The mRNA attaches to the ribosome at the mRNA's start codon. Then the ribosome slides along the mRNA molecule, reading one codon at a time. Molecules of tRNA ferry amino acids to the mRNA and add them to a growing protein chain. Translation stops when a stop codon is encountered. The protein chain then separates from the ribosome.
Mutations (pp. 441-442)
• A point mutation is a change in one or a few nucleotides in the sequence of a DNA molecule. When one nucleotide is mistakenly substituted for another, the function of the resulting protein may or may not affect the function of the protein. The insertion or deletion of a nucleotide always changes the resulting protein.
Regulating Gene Activity (p. 442)
• Gene activity is regulated at several levels. Usually, most of the DNA is folded and coiled. For a gene to be active, the region of DNA in which it is located must be uncoiled. Gene activity can be affected by other segments of DNA. Regions of DNA called enhancers can increase the amount of RNA produced. Chemical signals such as regulatory proteins or hormones can also affect gene activity.
Genetic Engineering (pp. 442-450)
• Genetic engineering is the purposeful manipulation of genetic material by humans. It can be used to produce large quantities of a particular gene product or to transfer a desirable genetic trait from one species to another or to another member of the same species.
• Genetic engineering uses restriction enzymes to cut the source DNA, which contains the gene of interest, and the vector DNA at specific places, creating sticky ends composed of unpaired complementary bases that allow the cut segments to recombine. The recombinant vector is then used to transfer the recombinant DNA to a host cell. Common vectors include bacterial plasmids and viruses. The host cell is often a type of cell that reproduces rapidly, such as a bacterium or yeast cell. Each time a host cell divides, both daughter cells receive a copy of the gene of interest. Introducing a gene from one species into a different species results in a transgenic plant or animal.
• Genetic engineering has had many applications in plant and animal agriculture, environmental science, and medicine.
• In gene therapy, a healthy form of a gene is introduced into body cells to correct problems caused by a defective gene.
Genomics (pp. 450-451)
• A genome consists of all the genes in a single organism.
Genomics is the study of genomes and the interaction between genes and the environment. It is now believed that the human genome consists of 20,000 to 25,000 genes.
• Scientists use microarrays to analyze gene activity under different conditions. It uses DNA chips—glass slides with thousands of DNA segments stamped on them. The information may be helpful in treating genetic diseases. Microarray analysis also allows scientists to discover small differences in the gene sequences of people. Scientists hope to use this information to develop individualized treatments.
• Large portions of our DNA are the same as the DNA in other organisms. The closer the evolutionary relationship, the greater portion of DNA we have in common.
Reviewing the Concepts
1. Describe the structure of DNA. p. 434
2. Explain why complementary base pairing is crucial to exact replication of DNA. p. 435
3. Why is DNA replication described as semiconservative? p. 436
4. Explain the roles of transcription and translation in converting the DNA message to a protein. pp. 436-437
5. In what ways does RNA differ from DNA? pp. 436-437
6. What roles do mRNA, tRNA, and rRNA play in the synthesis of protein? p. 437
7. Define codon. What role do codons play in protein synthesis? p. 438
8. What is the role of an anticodon? pp. 438-439
9. Describe the events that occur during the initiation of protein synthesis, the elongation of the protein chain, and the termination of synthesis. pp. 439-441
10. Why does a deletion have such a major effect on a cell? pp. 441-442
11. How is gene activity regulated? p. 442
12. Define genetic engineering. Explain the roles of restriction enzymes and vectors in genetic engineering. pp. 442-445
13. Describe some of the ways in which genetic engineering has been used in farming and medicine. pp. 445-450
14. What is gene therapy? pp. 448-450
15. The complementary base for thymine is
a. adenine.
b. cytosine.
c. guanine.
d. uracil.
16. Although the amount of any particular base in DNA will vary among individuals, the amount of guanine will always equal the amount of
a. thymine.
b. adenine.
c. cytosine.
d. uracil.
17. A codon is located on
a. DNA.
b. mRNA.
c. tRNA.
d. rRNA.
18. Translation produces
a. a polypeptide chain.
b. mRNA complementary to a template strand of DNA.
c. tRNA complementary to mRNA.
d. rRNA.
19. The anticodon is located on a molecule of _____.
20. In RNA, the nucleotide _____ binds with adenine.
21. In genetic engineering, the staggered cuts in DNA that allow genes to be spliced together are made by ____.
Applying the Concepts
Use the genetic code in Figure 21.5 (p. 438) to answer questions 1 and 2.
1. What would be the amino acid sequence in the polypeptide resulting from a strand of mRNA with the following base sequence?
AUG ACA UAU GAG ACG ACU
2. The following are base sequences in four mRNA strands: one normal and three with mutations. (Keep in mind that translation begins with a start codon and ends with a stop codon.) Which of the mutated sequences is likely to have the most severe effects? Why? Which would have the least severe effects? Why?
Normal mRNA: AUG ACA UAU GAG ACG ACU
Mutation 1: AUG ACC UAC GAA ACG ACC
Mutation 2: AUG ACU UAA GAG ACG ACA
Mutation 3: AUG ACG UAU GAG ACG ACG
3. You are a crime scene investigator testifying at a murder trial. The DNA fingerprints shown on the right are those of a bloodstain at the murder scene (not the victim's blood) and those of seven suspects (numbered 1 through 7). Which suspect's blood matches the bloodstain from the crime scene?
Becoming Information Literate
Describe some of the possible medical uses of information gained from the Human Genome Project. What ethical issues are raised by the project? Use at least three reliable sources (journals, books, websites) to answer these questions. Explain why you chose those sources.
___________________________________________________
1 We will develop this concept as the chapter proceeds. Some genes code for a polypeptide that is only part of a functional protein. A gene can also code for RNA that forms part of a ribosome or that transports amino acids during protein synthesis.
2 Plasmids seem to have evolved as a means of moving genes between bacteria. A plasmid can replicate itself and pass, with its genes, into another bacterium.