Medical Microbiology
Section 1 The adversaries – microbes
3 The viruses
Introduction
Viruses differ from all other infectious organisms in their structure and biology, particularly in their reproduction. Although viruses carry conventional genetic information in their DNA or RNA, they lack the synthetic machinery necessary for this information to be processed into new virus material. Viruses are metabolically inert and can replicate only after infecting a host cell and parasitizing the host’s ability to transcribe and/or translate genetic information. Viruses infect every form of life. They cause some of the most common and many of the most serious diseases of humans. Some insert their genetic material into the human genome and can cause cancer. Others have the ability to remain latent in different cell types and then reactivate at any time but especially if the body is stressed. Viruses are difficult targets for antiviral agents as it is difficult to target only those cells infected by the virus. However, many can be controlled by vaccines.
Viruses share some common structural features
Viruses range from very small (poliovirus, at 30 nm) to quite large – vaccinia virus, at 400 nm, is as big as small bacteria). Their organization varies considerably between the different groups, but there are some general characteristics common to all:
• The genetic material, in the form of single-stranded (ss) or double-stranded (ds), linear or circular RNA or DNA, is contained within a coat or capsid, made up of a number of individual protein molecules (capsomeres).
• The complete unit of nucleic acid and capsid is called the ‘nucleocapsid’, and often has a distinctive symmetry depending upon the ways in which the individual capsomeres are assembled (Fig. 3.1). Symmetry can be icosahedral, helical or complex.
• In many cases, the entire virus particle or ‘virion’ consists only of a nucleocapsid. In others, the virion consists of the nucleocapsid surrounded by an outer envelope or membrane (Fig. 3.2). This is generally a lipid bilayer of host cell origin, into which virus proteins and glycoproteins are inserted.
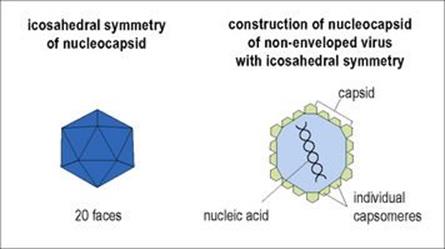
Figure 3.1 Symmetry and construction of the viral nucleocapsid.
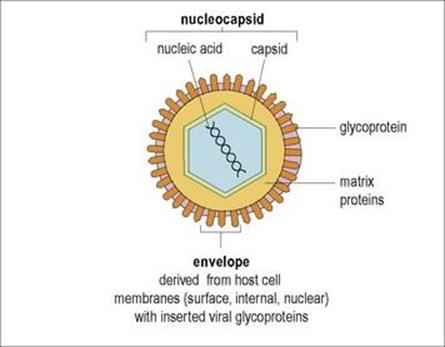
Figure 3.2 Construction of an enveloped virus.
The outer surface of the virus particle is the part that first makes contact with the membrane of the host cell
The structure and properties of the outer surface of the virus particle are therefore of vital importance in understanding the process of infection. In general, naked (envelope-free) viruses are resistant and survive well in the outside world; they may also be acid and bile-resistant, allowing infection through the gastrointestinal tract. Enveloped viruses are more susceptible to environmental factors such as drying, gastric acidity and bile. These differences in susceptibility influence the ways in which these viruses can be transmitted.
Infection of host cells
The stages involved in infection of host cells are summarized in Figure 3.3 (see also Fig. 2.6).
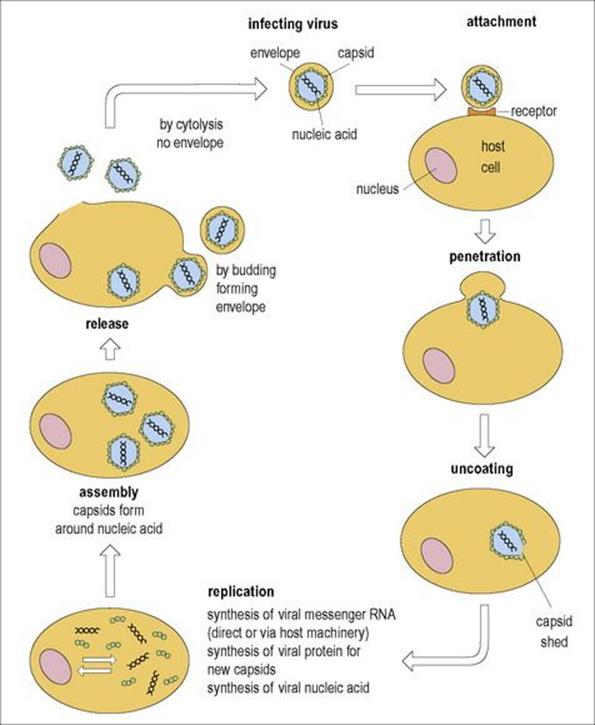
Figure 3.3 Stages in the infection of a host’s cell and replication of a virus. Several thousand virus particles may be formed from each cell.
Virus particles enter the body of the host in many ways
The most common forms of virus transmission (Fig. 3.4; see Ch. 13) are:
• via inhaled droplets (e.g. rhinovirus, influenza viruses)
• in food or water (e.g. hepatitis A virus, noroviruses)
• by direct transfer from other infected hosts (e.g. HIV, hepatitis B virus)
• from bites of vector arthropods (e.g. yellow fever virus, West Nile virus).
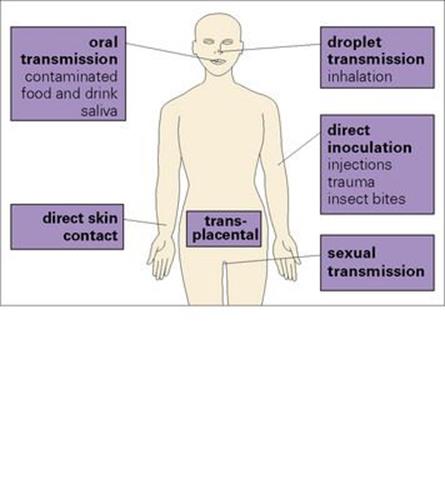
Figure 3.4 Routes by which viruses enter the body.
Viruses show host specificity and usually infect only one or a restricted range of host species. The initial basis of specificity is the ability of the virus particle to attach to the host cell
The process of attachment to, or adsorption by, a host cell depends on general intermolecular forces, then on more specific interactions between the molecules of the nucleocapsid (in naked viruses) or the virus membrane (in enveloped viruses) and the molecules of the host cell membrane. In many cases, there is a specific interaction with a particular host molecule, which therefore acts as a receptor. Influenza virus, for example, attaches by its haemagglutinin to a glycoprotein (sialic acid) found on cells of mucous membranes and on red blood cells; other examples are given inTable 3.1. Attachment to the receptor is followed by entry into the host cell.
Table 3.1 Viruses may use more than one receptor to gain entry into the host cell
|
Cell membrane receptors for virus attachment |
|
|
Virus |
Receptor molecule |
|
Influenza |
Sialic acid receptor on lung epithelial cells and upper respiratory tract |
|
Rabies |
Acetylcholine receptor |
|
HIV |
CD4: Primary receptor |
|
Epstein–Barr virus |
C3d receptor on B cells |
|
Human parvovirus B19 |
P antigen on erythoid progenitor cells |
|
Hepatitis C virus |
Epidermal growth factor receptor and ephrin receptor A2 are host co-factors for viral entry |
|
Human rhinoviruses |
Divided into two groups based on receptor binding: |
Once in the host’s cytoplasm the virus is no longer infective
After fusion of viral and host membranes, or uptake into a phagosome, the virus particle is carried into the cytoplasm across the plasma membrane. At this stage, the envelope and/or the capsid are shed and the viral nucleic acid released. The virus is now no longer infective: this ‘eclipse phase’ persists until new complete virus particles reform after replication. The way in which replication occurs is determined by the nature of the nucleic acid concerned.
Replication
Viruses must first synthesize messenger RNA (mRNA)
Viruses contain either DNA or RNA, never both. The nucleic acids are present as single or double strands in a linear (DNA or RNA) or circular (DNA) form. The viral genome may be carried on a single molecule of nucleic acid or on several molecules. With these options, it is not surprising that the process of replication in the host cell is also diverse. In viruses containing DNA, mRNA can be formed using the host’s own RNA polymerase to transcribe directly from the viral DNA. The RNA of viruses cannot be transcribed in this way, as host polymerases do not work from RNA. If transcription is necessary, the virus must provide its own polymerases. These may be carried in the nucleocapsid or may be synthesized after infection.
RNA viruses produce mRNA by several different routes
In dsRNA viruses, one strand is first transcribed by viral polymerase into mRNA (Fig. 3.5). In ssRNA viruses, there are three distinct routes to the formation of mRNA:
1. Where the single strand has the positive (+) sense configuration (i.e. has the same base sequence as that required for translation), it can be used directly as mRNA.
2. Where the strand has the negative (–) sense configuration, it must first be transcribed, using viral polymerase, into a positive sense strand, which can then act as mRNA.
3. Retroviruses follow a completely different route. Their positive sense ssRNA is first made into a negative sense ssDNA, using the viral reverse transcriptase enzyme carried in the nucleocapsid, and dsDNA is then formed which enters the nucleus and becomes integrated into the host genome. This integrated viral DNA is then transcribed by host polymerase into mRNA.
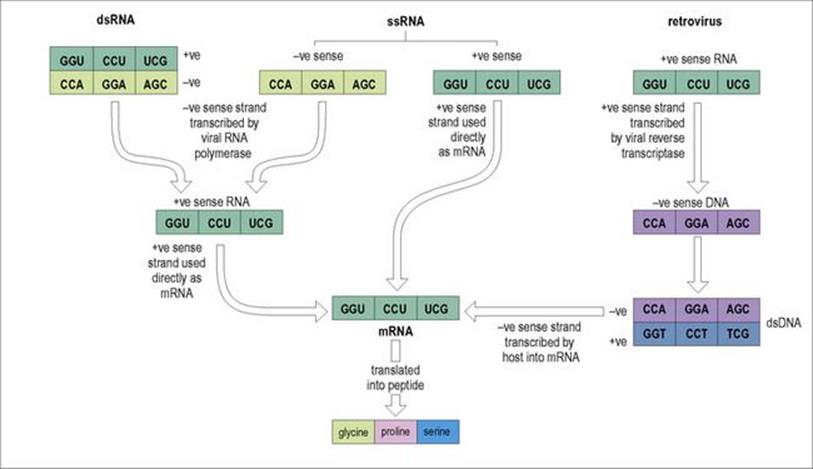
Figure 3.5 Ways in which genomic RNA of RNA viruses can be transcribed into messenger RNA (mRNA) before translation into proteins. + ve, positive sense; − ve, negative sense; ds, double stranded; ss, single stranded.
Viral mRNA is then translated in the host cytoplasm to produce viral proteins
Once viral mRNA has been formed, it is translated using host ribosomes to synthesize viral proteins (Fig. 3.6). Viral mRNA, which is usually ‘monocistronic’ (i.e. has a single coding region) can displace host mRNA from ribosomes so that viral products are synthesized preferentially. In the early phase, the proteins produced (enzymes, regulatory molecules) are those that will allow subsequent replication of viral nucleic acids; in the later phase, the proteins necessary for capsid formation are produced.
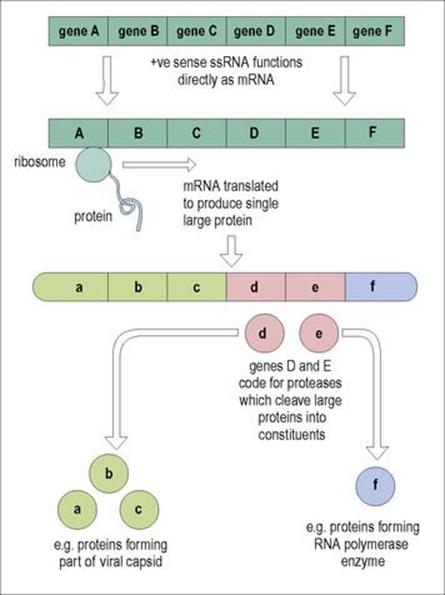
Figure 3.6 Translation and cleavage of viral proteins from messenger RNA (mRNA). + ve, positive sense; ss, single stranded.
In viruses where the genome is a single nucleic acid molecule, translation produces a large multifunctional protein, a polyprotein, which is then cleaved enzymatically to produce a number of distinct proteins. In viruses where the genome is distributed over a number of molecules, several mRNAs are produced, each being translated into separate proteins. After translation, the proteins may be glycosylated, again using host enzymes.
Viruses must also replicate their nucleic acid
In addition to producing molecules for the formation of new capsids, the virus must replicate its nucleic acid to provide genetic material for packaging into these capsids. In positive sense ssRNA viruses such as poliovirus, a polymerase translated from viral mRNA produces negative sense RNA from the positive sense template, which is then transcribed repeatedly into more positive strands. Further cycles of transcription then occur, resulting in the production of very large numbers of positive strands, which are packaged into new particles using structural proteins translated earlier from mRNA (Fig. 3.7).
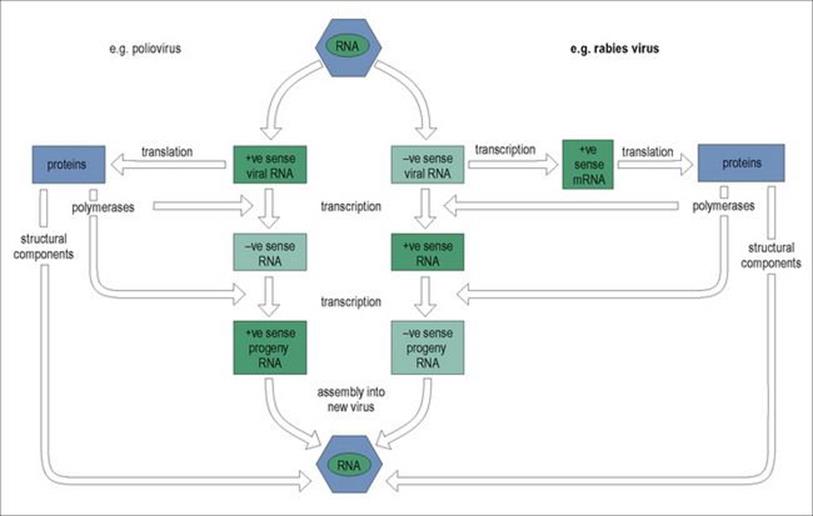
Figure 3.7 The ways in which genomic RNA of RNA viruses is replicated. + ve, positive sense; –ve, negative sense; mRNA, messenger RNA.
In negative sense ssRNA viruses (e.g. rabies virus), transcription by viral polymerase produces positive sense RNA strands from which new negative sense RNA is produced (Fig. 3.7). In the rabies virus, this replication occurs in the host cell cytoplasm, but in others (e.g. measles and influenza virus) replication takes place within the nucleus, large numbers of negative sense RNA molecules being transcribed for new particles.
Nucleic acid replication follows a similar pattern in dsRNA viruses (e.g. rotavirus) in that positive sense RNA strands are produced. These then act as templates in a subviral particle for the synthesis of new negative sense strands to restore the double stranded condition.
Replication of viral DNA occurs in the host nucleus – except for poxviruses, where it takes place in the cytoplasm
Viral DNA may become complexed with host histones to produce stable structures. With herpesviruses, mRNA translated in the cytoplasm produces a DNA polymerase that is necessary for the synthesis of new viral DNA; adenoviruses use both viral and host enzymes for this purpose. With retroviruses (e.g. HIV), synthesis of new viral RNA occurs in the nucleus, host RNA polymerase transcribing from the viral DNA that has become integrated into the host genome (see Fig. 3.5). Hepatitis B virus, a partially dsDNA virus, is unique in using a ssRNA intermediate transcribed from its DNA in order to synthesize new DNA. Retroviruses and hepatitis B are the only viruses affecting humans that have reverse transcriptase activity.
The final stage of replication is assembly and release of new virus particles
Assembly of virus particles involves the association of replicated nucleic acid with newly synthesized capsomeres to form a new nucleocapsid. This may take place in the cytoplasm or in the nucleus of the host cell. Enveloped viruses go through a further stage before release. Envelope proteins and glycoproteins, translated from viral mRNA, are inserted into areas of the host cell membrane (usually the plasma membrane). The progeny nucleocapsids associate specifically with the membrane in these areas, via the glycoproteins, and bud through it (Fig. 3.8). The new virus acquires the host cell membrane plus viral molecules as an outer envelope, and viral enzymes, such as the neuraminidase of influenza virus, may assist in this process (see details for influenza virus in Ch. 19). Host enzymes (e.g. cellular proteases) may cleave the initial large envelope proteins, a process that is necessary if the progeny viruses are to be fully infectious. In herpesviruses, acquisition of a membrane occurs as the nucleocapsids bud from the inner nuclear membrane. Release of enveloped viruses can occur without causing cell death so that infected cells continue to shed virus particles for long periods.
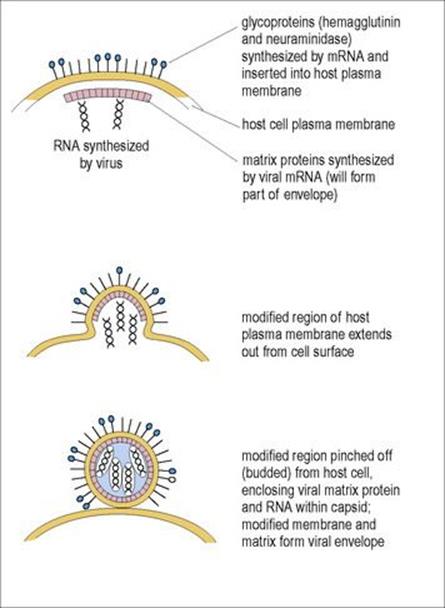
Figure 3.8 Release of enveloped RNA virus by budding through host cell membrane. Influenza A virus is shown in this example.
Insertion of viral molecules into the host cell membrane results in the host cell becoming antigenically different. Expression of viral antigens in this way is a major factor in the development of antiviral immune responses.
Outcome of viral infection
Viral infections may cause cell lysis or be persistent or latent
In lytic infections, the virus goes through a cycle of replication, producing many new virus particles. These are released by cell lysis. This host cell destruction is the typical consequence of infection with polio or influenza viruses. With other infections, such as hepatitis B, the cell may remain alive and continue to release virus particles at a slow rate. These ‘persistent’ infections are of great epidemiologic importance, as the infected person may act as a symptomless carrier of the virus, providing a continuing source of infection (see Ch. 16). In both lytic and persistent infections, the virus undergoes replication. However, in latent infections, the virus remains quiescent, and the genetic material of the virus may:
• exist in the host cell cytoplasm (e.g. herpesvirus)
• be incorporated into the genome (retroviruses, hepatitis B).
Replication does not take place until some signal triggers a release from latency. The stimuli that result in release are not fully understood in all cases. In herpes simplex infection, stress can activate the virus, resulting in an active infection seen as cold sores.
Some viruses can ‘transform’ the host cell into a tumour or cancer cell
Lytic, persistent and latent infections involve essentially normal host cells, although cellular metabolic and regulatory processes can be severely disrupted. Some viruses, however, can ‘transform’ the host cell, malignant transformation being the change of a differentiated host cell into a tumour or cancer cell (see Ch. 17). Transformed cells show changes in morphology, behaviour and biochemistry. Controlled growth patterns and contact inhibition are lost, so that cells continue to divide and form random aggregations. They become invasive and can form tumours if injected into animals. However, not all transformed cells give rise to harmful tumours in vivo. Warts, for example, may be benign growths on the skin of the hands or feet caused by one group of papillomaviruses, or genital warts caused by a different group of specific papillomaviruses may lead to cervical cancer.
Cancer-inducing viruses are found in several different groups including both DNA and RNA viruses. They include the human T-cell lymphotropic virus type 1 (see below), the Epstein–Barr virus, papillomaviruses 16 and 18 and hepatitis B and C virus infections (see Ch. 17). Although the end results of transformation may be similar, the mechanisms involved vary between different viruses. However, all involve interference with the normal regulation of division and response to external growth-promoting and growth-inhibiting factors. These changes come about after viral nucleic acid is incorporated into the host genome. Finally, cancer is not always the result of some of these infections. Papillomaviruses are present in cervical cancer but additional cellular events are needed for most of the other viral infections to result in tumours.
For example, the Rous sarcoma virus is a retrovirus that causes cancer in chickens. 2011 was the hundredth anniversary of Francis Rous demonstrating that this chest tumour could be transmitted by giving tumour extracts that were cell-free to chickens related to the same brood. Transformation arises from the introduction into the host genome of a viral oncogene, v-src. This codes for an activated and overexpressed protein – tyrosine kinase, an enzyme involved in the phosphorylation of tyrosine residues in target proteins. This leads to some molecular events and changes in phenotype in transformed host cells and subsequent tumorigenesis as a result of the viral infection. A urokinase-type plasminogen activator (PLAU) gene is induced by v-src and highly up-regulated. PLAU is a protease enzyme that lyses fibrin and breaks down the extracellular matrix promoting cancer cell stickiness and spread
The first human tumour virus was discovered in 1964 when Epstein–Barr virus (EBV) was found by electron microscopic analysis of cells of a tumour called Burkitt’s lymphoma seen in African patients.
More than 20 retroviral oncogenes are now known (Table 3.2). Of the retrovirus family, the human T-cell lymphotropic virus (HTLV type 1) is a cancer-causing virus in humans despite neither possessing a viral oncogene nor directly activating a cellular oncogene (see below). In contrast, HIV type 1 and 2 virus infections compromise the host’s immune system, resulting in tumours associated with other viruses including EBV and Kaposi’s sarcoma herpesvirus (KSHV) also known as human herpesvirus 8 (HHV-8). A larger number of retroviruses cause cancers in animals.
Table 3.2 Oncogenes, gene products, viruses known to carry them and associated human and animal diseases
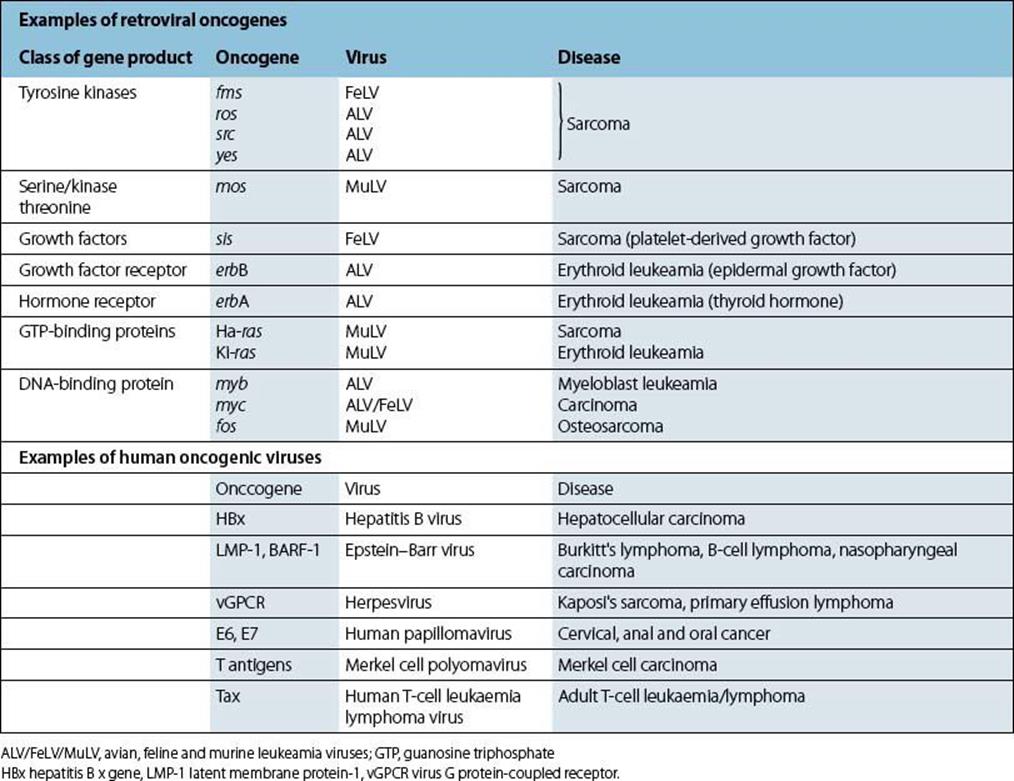
Tumour formation as a result of viral infection: direct and indirect mechanisms
Viruses associated with cancer may do so by direct means, by expressing viral oncogenes that transform the cell as mentioned above. Alternatively, they may do so indirectly by chronically infecting the cells resulting in inflammation and mutations that result in tumour formation. In addition, some virus infections including hepatitis B and C may produce various proteins that start oncogenic transformation of cells. New mechanisms are being detected by knocking out the action of certain genes and comparing the results with the control group.
Viral oncogenes have probably arisen from incorporation of host oncogenes into the viral genome during viral replication
Oncogenes are designated by short acronyms, preceded by ‘v’ if a viral oncogene is described (e.g. v-myc) or by ‘c’ for a cellular (host) oncogene (e.g. c-myc). DNA probes made from copies of the Rous sarcoma virus src oncogene have revealed complementary DNA in both infected and normal chicken cells, as well as in cancerous and normal human cells. This striking finding has since been repeated with many other retroviral oncogene sequences and it is now known that these can make up as much as 0.03–0.3% of the mammalian genome. Oncogene sequences have been identified in a wide variety of animals, from man to fruit flies, implying that they are conserved because of some valuable function. Which came first, host or viral oncogenes? The fact that host oncogenes contain introns, whereas viral oncogenes do not, and that their chromosomal positions are fixed, implies that they, and not the viral forms, are the original genes.
From what we now know about the gene products of viral oncogenes we can guess that cellular oncogenes (or ‘proto-oncogenes’) probably play an important role in host cell growth regulation. They may code for growth factors themselves, for cell surface receptor molecules that bind specific growth factors, for components of intracellular signalling systems, or for DNA-binding proteins that act as transcription factors.
The Rous sarcoma virus src oncogene is incorporated within the viral genome adjacent to the gene coding for viral envelope proteins (Fig. 3.9). Unlike other strongly transforming viruses, the Rous virus has all three genes (gag, pol and env) necessary for replication. In the others, termed ‘defective’ transforming viruses, incorporation of an oncogene results in deletion of genetic material in the regions coding for the pol and/or env genes, so preventing replication. This becomes possible only with help from genetically complete helper viruses.
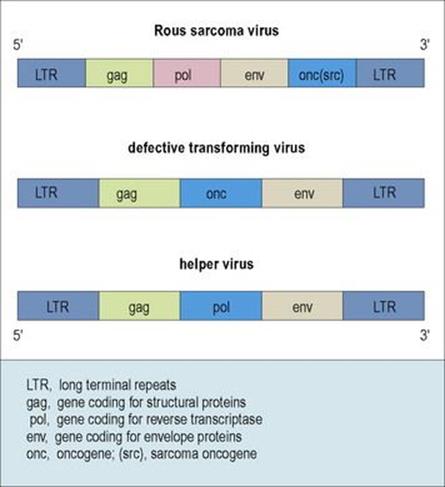
Figure 3.9 Rous sarcoma virus can transform the host cell and replicate because it has both the oncogene src and a complete genome. Some transforming viruses are defective – they carry the oncogene, but lack genes for full replication. Helper viruses can supply these genes.
Oncogenes can be carried from one cell to another within the same host or from one host to another. This can occur through ‘vertical’ transmission, from mother to offspring, through passage of viruses in gametes, across the placenta or in milk. It can also occur by ‘horizontal’ transmission, the virus passing in, for example, saliva or urine (see Ch. 13).
Transformation of a cell occurs:
• when viral oncogenes are incorporated into the host genome (as in Rous sarcoma virus)
• when viral DNA is inserted near to a cellular oncogene.
The former may be due to mutations in the oncogene sequence while in the viral genome; single base changes in cellular oncogenes are known to confer the ability to transform normal cells. The latter may reflect altered expression of the host oncogene through disturbance of normal regulatory influences. Altered expression can occur whether the insertion is of a retroviral oncogene or of non-oncogenic viral DNA; it can also occur as a result of exposure to a variety of carcinogens. The products of cellular oncogenes are normally used in series to regulate cellular proliferation in a carefully controlled manner. Viral oncogene products or overexpressed cellular oncogene products short circuit and overload this complex control system, resulting in unregulated cell division.
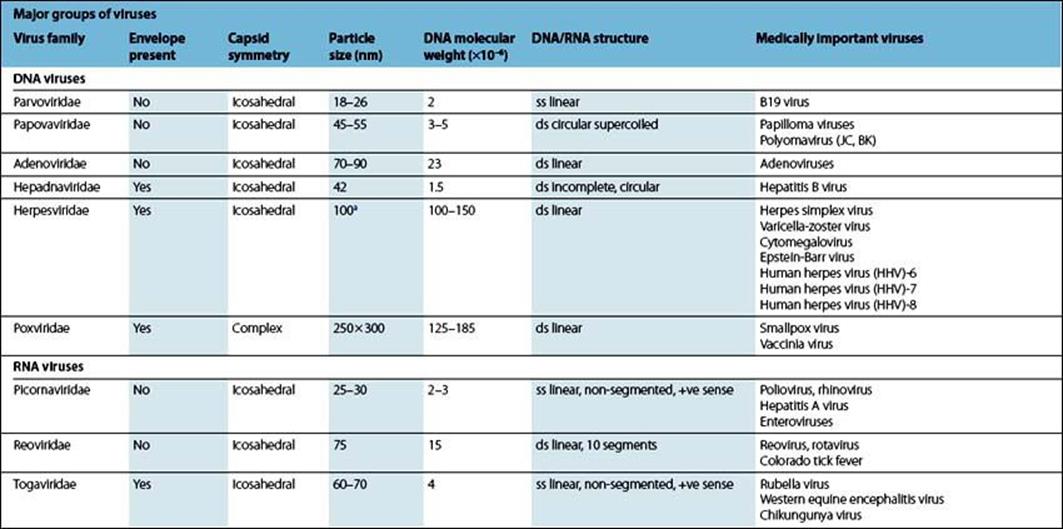
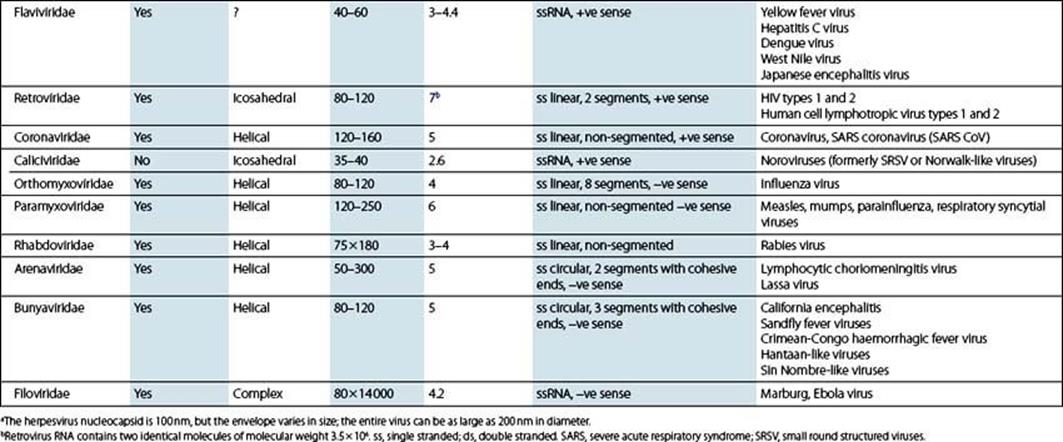
Major groups of viruses
The classification of viruses into major groups (families) is based on a few simple criteria (Table 3.3 and the Pathogen Parade). These include:
• the type of nucleic acid in the genome
• the number of nucleic acid strands and their polarity
• the mode of replication
• the size, structure and symmetry of the virus particle.
Table 3.3 Summary of major families of viruses
![]()
 Key Facts
Key Facts
• Viruses have RNA or DNA but absolutely depend on the host to process their genetic information into new virus particles.
• The outer surface of a virus (capsid or envelope) is essential for host cell contact and entry, and determines the capacity to survive in the outside world.
• Viruses are most often transmitted in droplets, in food and water or by intimate contact.
• Replication of viral RNA or DNA is a complex process, making use of host and/or viral enzymes.
• RNA of retroviruses becomes integrated into the host genome.
• New virus particles are released by cell lysis or by budding through the host cell membrane.
• Some viruses, such as herpesviruses, may become latent and require a trigger to resume replication; others replicate at a slow rate, persisting as a source of infection in symptomless carriers.
• A number of viruses transform the host cell, by interfering with normal cellular regulation, resulting in the development of a cancer cell. This may be the result of the activity of viral or cellular oncogenes.
![]()
![]()
 Conflicts
Conflicts
Viruses have developed a cunning strategy as hardy infectious agents as, once they have infected the host cell, they may lie latent or integrate within the host cell chromosome and reactivate, potentially transmitting the infection to others. The host may not be too incapacitated, ensuring they can infect those susceptible. In addition, the host has to have a full immunosurveillance repertoire to suppress all these viruses waiting to step up to the plate. Once the defences are lowered by stress, immunosuppression or trauma, for example, active viral replication can occur.
Viruses may have a number of options with respect to receptors they can attach to and subsequently infect the host. They may be able to cross species barriers as well and not affect the reservoir host. With respect to transmissibility, their job description includes the ability to exist in blood and other body fluids, be aerosolized and to be carried by insect vectors.
To keep the host’s immune system on its toes, most of the RNA viruses can subtly change their genetic make-up and drift away from the circulating strain, thus evading the immune response. Alternatively, they may have a number of genotypes with a different susceptibility to antiviral agents, are not cross-protective therefore ensuring a multivalent vaccine is required as a preventative measure, and are associated with a different clinical illness spectrum.
Viruses make full use of the cellular replicative machinery and therefore an antiviral agent has difficulty targeting the virus without affecting the host cell. As a result, most antiviral agents can adversely affect the host. This means that individuals taking certain antiviral agents have to be monitored carefully, as treatment can potentially lead to side effects including bone marrow suppression, renal toxicity and mitochondrial disorders.
What can the host do to offset all these advantages? Antiviral vaccines have been a major success, behavioural changes can limit the chances of infection and, increasingly, more precise chemotherapeutic targets are being identified.
![]()