CONCEPTS IN BIOLOGY
PART III. MOLECULAR BIOLOGY, CELL DIVISION, AND GENETICS
8. DNA and RNA. The Molecular Basis of Heredity
8.4. Protein Synthesis
DNA and RNA are both important in the protein-making process. In the cell, the DNA nucleotides are used as a genetic alphabet, arranged in sets of three (e.g., ATC, GGA, TCA, CCC) to form code words in the DNA language. It is the sequence of these code words in DNA that dictates which amino acids are used, and the order in which they appear in a protein. DNA molecules are very long and code for many proteins along their length. Proteins are synthesized in two steps; transcription and translation.
Step One: Transcription—Making RNA
Transcription is the process of using DNA as a template (stencil) to synthesize RNA. The enzyme RNA polymerase “reads” the sequence of DNA nitrogenous bases and follows the base-pairing rules between DNA and RNA to build the new RNA molecule (figure 8.6a-c). RNA polymerase attaches to the DNA and moves along the grooves scanning for base sequences that act as markers, or signs, that a gene is nearby. The enzyme looks for a promoter sequence (figure 8.6d). This is a specific sequence of DNA nucleotides that indicates the location of a protein-coding region and identifies which of the two DNA strands should be used. The coding strand of DNA is the side that serves as a template for the synthesis of RNA. The strand of DNA that is not read directly by the enzymes is the non-coding strand. Without promoter sequences, RNA polymerase will not transcribe the gene.
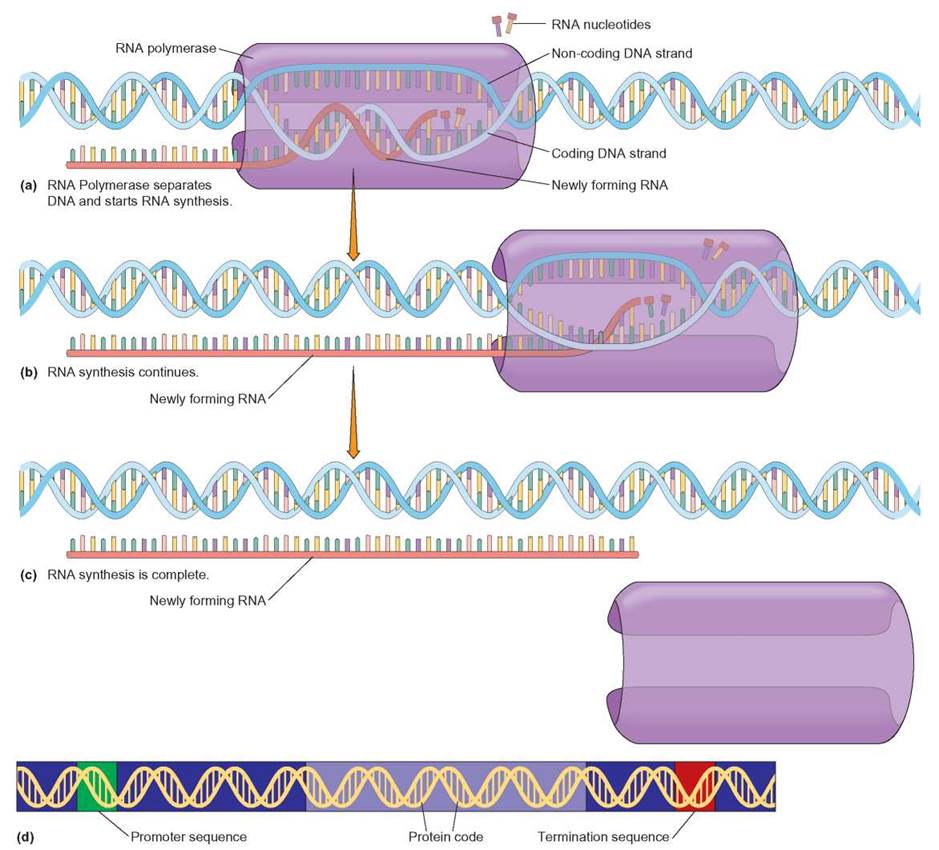
FIGURE 8.6. Transcription of DNA to RNA
This figure illustrates the basic events that occur during transcription. (a) An enzyme, RNA polymerase, attaches to the DNA at the promoter sequence (see d) and then separates the complementary strands. The enzyme then proceeds along the DNA strand in the correct direction to find the protein coding region (see d) of the gene. (b) As RNA polymerase moves down the coding strand, new complementary RNA nucleotides are base-paired to one of the exposed DNA strands. The base-paired RNA nucleotides are linked together by RNA polymerase to form a new RNA molecule that is complementary to the nucleotide sequence of the DNA. The termination sequence (see d) signals the RNA polymerase to end mRNA transcription, so that the RNA can leave the nucleus to aid in translation. (c) The newly formed (transcribed) RNA is then separated from the DNA molecule and used by the cell.
Transcription begins when the enzyme separates the two strands of the double-stranded DNA. Separating the two strands exposes their nitrogenous bases, so that the coding strand can be “read.” Reading is accomplished by bringing in new RNA nucleotides and base-pairing them with hydrogen bonds one at a time with the exposed DNA nucleotides. Once a match is made, the newly arrived RNA nucleotide is bonded in place by forming a covalent bond between the sugar of one RNA nucleotide and the phosphate of the next.
RNA polymerase stops transcribing the DNA when it reaches a termination sequence. Termination sequences are DNA nucleotide sequences that indicate when RNA polymerase should finish making an RNA molecule. Only one of the two strands of DNA is read to create a single strand of RNA for each gene.
There are several types of RNA: however, the three types we will focus on are messenger RNA (mRNA), transfer RNA (tRNA), and ribosomal RNA (rRNA). Each type of RNA is assembled in the nucleus from combinations of the same 4 nucleotides. However, each type of RNA has a distinct function in the process of protein synthesis that takes place in the cytoplasm. Messenger RNA (mRNA) carries the blueprint for making the necessary protein. Transfer RNA (tRNA) and ribosomal RNA (rRNA) are used in different ways to read the mRNA and bring the necessary amino acids together for assembly into a protein.
Step Two: Translation—Making Protein
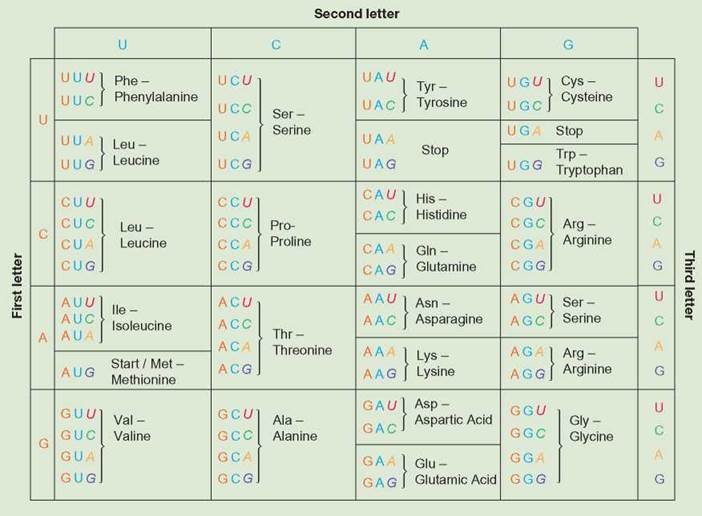
Translation is the process of using the information in RNA to direct protein synthesis by attaching amino acids to one another. The mRNA is read linearly in sets of three nucleotides called codons. A codon is a set of three nucleotides that codes for the placement of a specific amino acid. In the context of an mRNA molecule, the codon determines which amino acid should be added next to the protein during translation. Table 8.2 shows the mRNA nucleotide combinations of each codon and the corresponding amino acid. For example, the codon UUU corresponds to only the amino acid phenylalanine (Phe). There are 64 possible codons and only 20 commonly used amino acids, so there are multiple ways to code for many amino acids. For example, the codons UCU, UCC, UCA, and UCG all code for serine.
TABLE 8.2. Amino Acid-mRNA Dictionary and the 20 Common Amino Acids and Their Abbreviations
These are the 20 common amino acids used in the protein synthesis operation of a cell. Each has a known chemical structure and is coded for by specific mRNA codons.
Recall from chapter 4 that a ribosome is a nonmembranous organelle that synthesizes proteins. A ribosome is made of proteins and a type of RNA called ribosomal RNA (rRNA). Ribosomes usually exist in the cell as two pieces or subunits. There is a large subunit and a small subunit. During translation, the two subunits combine and hold the mRNA between them. With the mRNA firmly sandwiched into the ribosome, the mRNA’s codons are read and protein synthesis begins.
The cell has many ribosomes available for protein synthesis. Any of the ribosomes can read any of the mRNAs that come from the cell’s nucleus after transcription. Some ribosomes are free in the cytoplasm, whereas others are attached to the cell’s rough endoplasmic reticulum (ER). Proteins destined to be part of the cell membrane or packaged for release from the cell are synthesized on ribosomes attached to the endoplasmic reticulum. Proteins that are to perform their function in the cytoplasm are synthesized on ribosomes that are not attached to the endoplasmic reticulum. The process of translation can be broken down into three basic steps: (1) initiation, (2) elongation, and (3) termination.
Initiation
Protein synthesis begins with the small ribosomal subunit binding to a specific signal sequence of codons on the mRNA. The small ribosomal subunit moves along the mRNA and stops at the first AUG codon on the length of the RNA. This AUG codon is where translation begins. If an AUG is not found, translation does not occur. At the first AUG codon, the first amino acid (methionine, or MET) is positioned on the mRNA.
Amino acids are taken to the mRNA-ribosome complex by transfer RNA. Transfer RNA (tRNA) is responsible for matching the correct amino acid to the codons found in the mRNA nucleotide sequence (figure 8.7a). The cell’s tRNAs are able to match amino acids to the mRNA codons because of base pairing. The portion of the tRNA that interacts with mRNA is called the anticodon. The anticodon of tRNA is a short sequence of nucleotides that base-pairs with the nucleotides in the mRNA molecule. The other end of the tRNA carries an amino acid. The correct match between tRNAs and amino acids is made by an enzyme in the cell.
The start codon, AUG, is the first codon that is read in the mRNA to make any protein. Since the tRNA that binds to the AUG codon carries the amino acid methionine, the first amino acid of every protein is methionine (figure 8.7). If this first methionine is not needed for proper function of the protein, it can be later clipped off of the protein. After the methionine- tRNA molecule is lined up over the start codon, the large subunit of the ribosome joins the small subunit to bind the mRNA. When the two subunits are together, with the mRNA in the middle, the ribosome is fully formed. The process of forming the rest of the protein is ready to begin (figure 8.7).
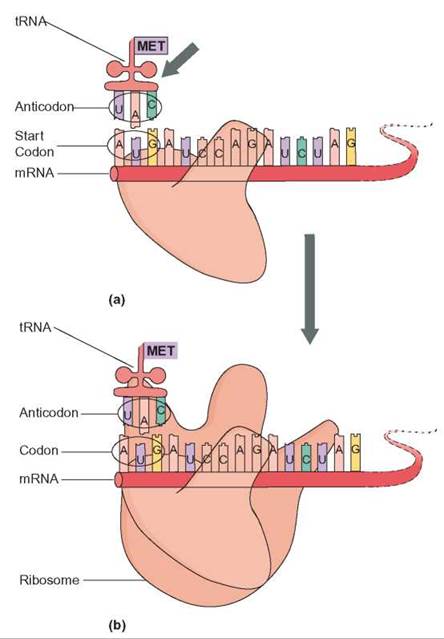
FIGURE 8.7. Initiation
(a) An mRNA molecule is positioned in the ribosome so that two codons are in position for transcription. The first of these two codons (AUG) is the initiation codon and is responsible for hydrogen bonding with the tRNA carrying the amino acid methionine (MET). The start tRNA aligns with the start codon. (b) The large subunit of the ribosome joins the small subunit. The ribosome is now assembled and able to translate the mRNA.
Elongation
Once protein synthesis is started, the ribosome coordinates a recurring series of events. Each time the ribosome works through this series of events, a new amino acid is added to the growing protein. In this way, a ribosome is like an assembly line that organizes the steps of a complicated assembly process. For each new amino acid, a new tRNA arrives at the ribosome with its particular amino acid. The ribosome adds the new amino acid to the growing protein (figure 8.8).
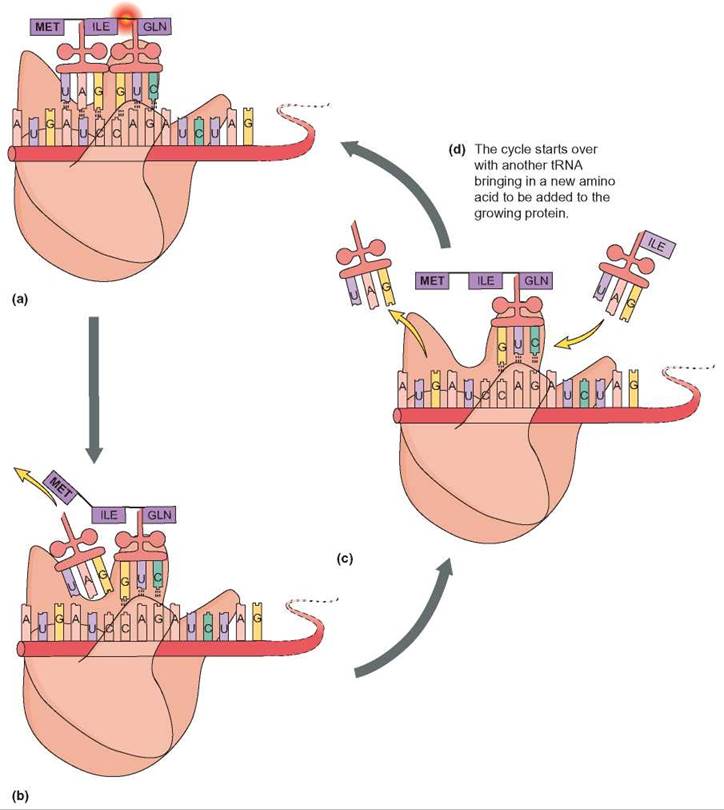
FIGURE 8.8. Elongation
(a) The two tRNAs align the amino acids isoleucine (ILE) and glutamine (GLN) so that they can be chemically attached to one another by forming a peptide bond. (b) Once the bond is formed, the first tRNA detaches from its position on the mRNA. (c) The ribosome moves down one codon on the mRNA. Another tRNA now aligns so that the next amino acid (ILE) can be added to the growing protein. (d) The process continues with a new tRNA, a new amino acid, and the formation of a new peptide bond.
Termination
The ribosome will continue to add one amino acid after another to the growing protein unless it encounters a stop signal (figure 8.9). The stop signal, in the mRNA, is also a codon. The stop codon can be either UAA, UAG, or UGA. When any of these three codons appears during the elongation process, a chemical release factor enters the ribosome. The release factor causes the ribosome to detach from the protein. When the protein releases, the ribosomal subunits separate and release the mRNA. The mRNA can be used to make another copy of the protein or can be broken down by the cell to prevent any more of the protein from being made. The two pieces of the ribosome can also be reused.
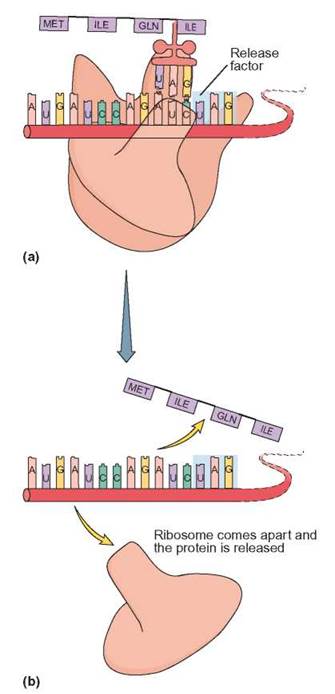
FIGURE 8.9. Termination
(a) A release factor will move into position over a termination codon—here, UAG. (b) The ribosome releases the completed amino acid chain. The ribosome disassembles and the mRNA can be used by another ribosome to synthesize another protein.
The Nearly Universal Genetic Code
The code for making protein from DNA is the same for nearly all cells. Bacteria, Archaea, algae, protozoa, plants, fungi and animals all use DNA to store their genetic information. They all transcribe the information in DNA to RNA. They all translate the RNA to synthesize protein using a ribosome. With very few exceptions, they all use the same three nucleotide codons to code for the same amino acid. In eukaryotic cells, transcription always occurs in the nucleus, and translation always occurs in the cytoplasm (figure 8.10).
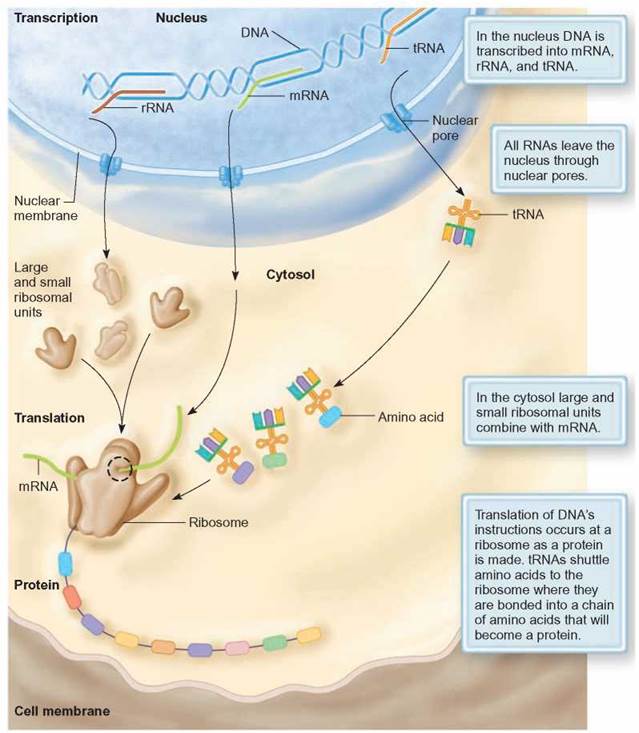
FIGURE 8.10. Summary of Eukaryotic Protein Synthesis
The genetic information in DNA is rewritten in the nucleus as RNA in the nucleus during transcription. The mRNA, tRNA, and rRNA move from the nucleus to the cytoplasm (cytosol), where the genetic information is read during translation by the ribosome.
The similarity of protein synthesis in all cells strongly argues for a common origin of all life forms. It also creates very exciting opportunities for biotechnology. It is now possible to synthesize human proteins, such as insulin, in bacteria, because bacteria and humans use the same code to make proteins. The production of insulin in this way can help create a cheap and plentiful source of medication for many of those who suffer from diabetes.
However, not all genetic information flows from DNA to RNA to proteins. Some viruses use RNA to store their genetic information. These viruses are called retroviruses. An example of a retrovirus is the human immunodeficiency virus, HIV. Retroviruses use their RNA to make DNA. This DNA is then used to transcribe more RNA. This RNA is then used to make proteins (Outlooks 8.1).
OUTLOOKS 8.1
Life in Reverse—Retroviruses
Acquired immunodeficiency syndrome (AIDS) is caused by a retrovirus called human immunodeficiency virus (HIV). HIV is a spherical virus that has RNA as its genetic material surrounded by a protein coat. In addition, the virus is surrounded by a phospholipid layer taken on from the cell's plasma membrane when the virus exits the host cell. When persons become infected with HIV, the outer phospholipid membrane of the virus fuses with the plasma membrane of the host cell and releases the virus with its RNA into the cell. In addition to its RNA genetic material, HIV carries a few enzymes; one is reverse transcriptase. When HIV enters a suitable host cell, the virus first uses reverse transcriptase to produce a DNA copy of its RNA. (This is the reverse of the normal process in cells that involves the enzyme transcriptase using DNA to make RNA.) Because this is the reverse (retro-) of what normally happens in a cell, RNA viruses are called retroviruses. The DNA produced by reverse transcriptase is spliced into the host cell's DNA. Only then does HIV become an active, disease-causing parasite. Once a DNA copy of the virus RNA is inserted into the host cell's DNA, the virally derived DNA is used to make copies of the viral RNA and its protein coat.
Understanding how HIV differs from DNA-based organisms has two important implications. First, the presence of reverse transcriptase in a human can be looked upon as an indication of retroviral infection because reverse transcriptase is not manufactured by human cells. However, because HIV is only one of several types of retroviruses, the presence of the enzyme in an individual does not necessarily indicate an HIV infection. It only indicates a type of retroviral infection. Second, antiviral drug treatments for HIV take advantage of vulnerable points in the retrovirus's life cycle. For example, interference with reverse transcriptase blocks the virus's ability to make DNA and lessens its chances of integrating into the host's DNA. This gives an infected person's body the opportunity to destroy the viruses and reduces the chance that the person will develop symptoms of the disease. Once cleared of viruses, the likelihood that the individual will transmit the virus to others is decreased.
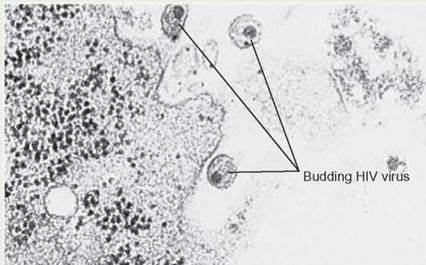
Budding HIV Virus
This electron micrograph shows HIV viruses leaving the cell. These viral particles can now infect another cell and continue the viral replication cycle unless medications prevent this from happening.
8.4. CONCEPT REVIEW
8. How does the manufacture of an RNA molecule differ from DNA replication?
9. If a DNA nucleotide sequence is TACAAAGCA, what is the mRNA nucleotide sequence that would base-pair with it?
10. What amino acids would occur in the protein chemically coded by the sequence of nucleotides in question 9?
11. How do tRNA, rRNA, and mRNA differ in function?
12. What are the differences among a promoter sequence, a termination sequence, and a release factor?
13. List the sequence of events that takes place when a DNA message is translated into protein.