CONCEPTS IN BIOLOGY
PART III. MOLECULAR BIOLOGY, CELL DIVISION, AND GENETICS
8. DNA and RNA. The Molecular Basis of Heredity
8.5. The Control of Protein Synthesis
Cells have many protein-coding sequences. Gene expression occurs when a cell transcribes and translates a gene. Cells do not make all their proteins at once. It would be a great waste of resources. Cells can control which genes are used to make proteins. In fact, the differences between the types of cells in the human body are due to the differences in the proteins produced. Cells use many ways to control gene expression in response to environmental conditions. Some methods help increase or decrease the amount of enzyme that is made by the cell. Other methods help change amino acid sequences to form a new version of the enzyme.
Controlling Protein Quantity
A cell process can be regulated by controlling how much of a specific enzyme is made. The cell regulates the amount of protein (enzymes are proteins) that is made by changing how much mRNA is available for translation into protein. The cell can use several strategies to control how much mRNA is transcribed.
DNA Packaging
The genetic material of humans and other eukaryotic organisms consists of strands of coiled, double-stranded DNA, which have histone proteins attached along its length. The histone proteins and DNA are not arranged randomly but, rather, come together in a highly organized pattern (figure 8.11). When packaged, the double-stranded DNA spirals around repeating clusters of eight histone spheres. Histone clusters with their encircling DNA are called nucleosomes. These coiled DNA strands with attached proteins become visible during cell division and are called nucleoproteins or chromatin fibers. Condensed like this, a chromatin fiber is referred to as a chromosome (Outlooks 8.2). The degree to which the chromatin is coiled provides a method for long-term control of protein expression. In tightly coiled chromatin, the promoter sequence of the gene is tightly bound so that RNA polymerase cannot attach and initiate transcription. Loosely packaged chromatin exposes the promoter sequence so that transcription can occur.
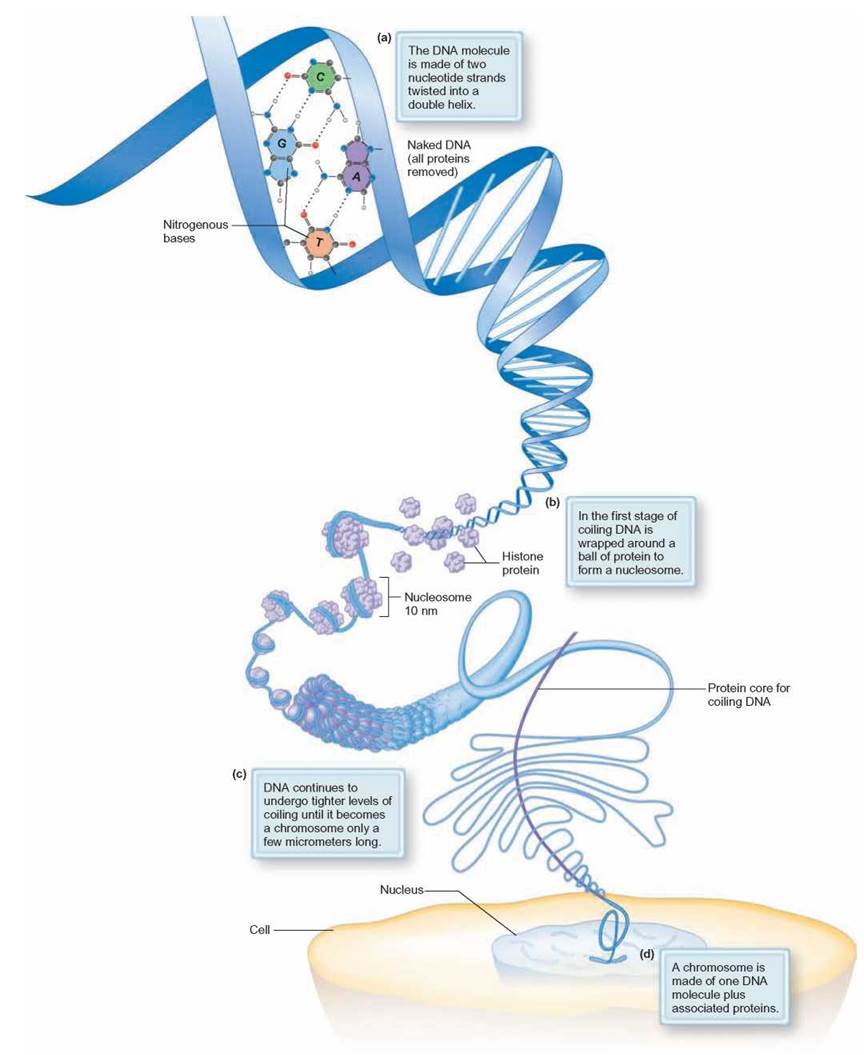
FIGURE 8.11. Eukaryotic Genome Packaging
During certain stages in the life cycle of a eukaryotic cell, the DNA is tightly coiled to form a chromosome. To form a chromosome, the DNA molecule is wrapped around a group of several histone proteins. Together, the histones and the DNA form a structure called the nucleosome. The nucleosomes are stacked together in coils to form a chromosome.
OUTLOOKS 8.2
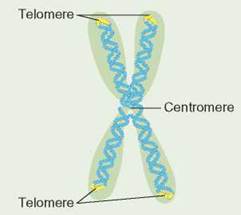
Telomeres
Each end of a chromosome contains a sequence of nucleotides called a telomere. In humans, these chromosome "caps" contain many copies of the following nucleotide base-pair sequence:
TTAGGG
AATCCC
Telomeres are very important segments of the chromosome. They are:
1. required for chromosome replication;
2. protect the chromosome from being destroyed by dangerous DNAase enzymes (enzymes that destroy DNA); and
3. keep chromosomes from bonding to one another end to end.
Evidence shows that the loss of telomeres is associated with cell "aging," whereas not removing them has been linked to cancer. Every time a cell reproduces itself, it loses some of its telomeres. However, in cells that have the enzyme telomerase, new telomeres are added to the ends of the chromosome each time the cells divide. Therefore, cells that have telomerase do not age as other cells do, and cancer cells are immortal because of this enzyme. Telomerase enables chromosomes to maintain, if not increase, the length of telomeres from one cell generation to the next.
The yellow regions on this drawing of a chromosome indicate where the telomeres are.
Enhancers and Silencers
Enhancer and silencer sequences are DNA sequences that act as binding sites for proteins. When proteins are bound to these sites, they affect the ability of RNA polymerase to transcribe a specific protein. Enhancer sequences increase protein synthesis by helping increase transcription. Silencer sequences decrease transcription. These DNA sequences are unique, because they do not need to be close to the promoter to function and they are not transcribed.
Transcription Factors
Transcription factors are proteins that control how available a promoter sequence is for transcription. The protein molecules bind to DNA around a gene’s promoter sequence and influence RNA polymerase’s ability to start transcription. There are many transcription factors in the cell. Eukaryotic transcription is so tightly regulated that transcription factors always guide RNA polymerase to the promoter sequence. A particular gene will not be expressed if its specific set of transcription factors is not available. Prokaryotic cells also use proteins to block or encourage transcription, but not to the extent that this strategy is used in eukaryotic cells.
RNA Degradation
Cells also regulate gene expression by limiting the length of time that mRNA is available for translation. Enzymes in the cell break down the mRNA, so that it can no longer be used to synthesize protein. The time that a given mRNA molecule lasts in a cell is dependant on the nucleotide sequences in the mRNA itself. These sequences are in areas of the mRNA that do not code for protein.
Controlling Protein Quality
Another way that cells can control gene expression is to change the amino acid sequences to form different versions of an enzyme. One of the most significant differences between prokaryotic and eukaryotic cells is that eukaryotic cells can make more than one type of protein from a single protein-coding region of the DNA. Eukaryotic cells are able to do this because the protein-coding regions of eukaryotic genes are organized differently than the genes found in prokaryotic (bacterial) cells. The fundamental difference is that the protein-coding regions in prokaryotes are continuous, whereas eukaryotic protein-coding regions are not. Many intervening sequences are scattered throughout the proteincoding sequence of genes in eukaryotic cells. These sequences, called introns, do not code for proteins. The remaining sequences, which are used to code for protein, are called exons. After the protein-coding region of a eukaryotic gene is transcribed into mRNA, the introns in the mRNA are cut out and the remaining exons are spliced together, end to end, to create a shorter version of the mRNA. It is this shorter version that is used during translation to produce a protein (figure 8.12).
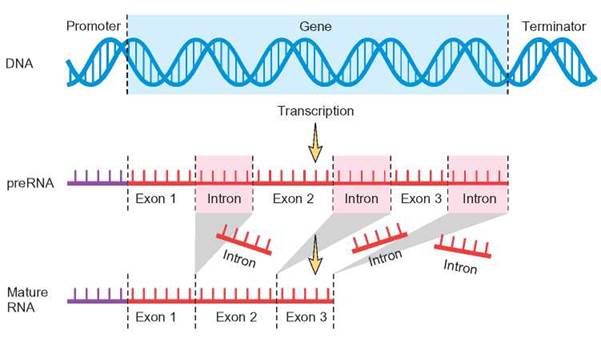
FIGURE 8.12. Transcription of mRNA in Eukaryotic Cells
This is a summary of the events that occur in the nucleus during the manufacture of mRNA in a eukaryotic cell. Notice that the original nucleotide sequence is first transcribed into an RNA molecule, which is later “clipped” and then rebonded to form a shorter version of the original. It is during this time that the introns are removed.
One advantage of having introns is that a single proteincoding region can make more than one protein. Scientists originally estimated that humans had 80,000 to 100,000 genes. This was based on techniques that allowed them to estimate the number of different proteins found in humans. When the human genome was mapped, scientists were surprised to find that humans have only about 25,000 genes. This suggests that many of our genes are capable of making several different proteins.
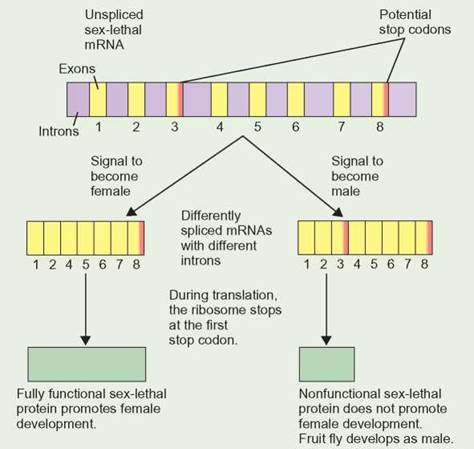
It is possible to make several different proteins from the same protein-coding region by using different combinations of exons. Alternative splicing is the process of selecting which exons will be retained during the normal process of splicing. Alternative splicing can be a very important part of gene regulation. One protein-coding region in fruit flies, sex-lethal, can be spliced into two different forms. One form creates a full-sized, functional protein. The other form creates a very small protein with no function. For the fruit fly, the difference between the two alternatively spliced forms of sex-lethal is the difference between becoming a male or becoming a female fruit fly (Outlooks 8.3).
OUTLOOKS 8.3
One Small Change—One Big Difference!
Male and female fruit flies produce the same unspliced mRNA from the sex-lethal gene. A cellular signal determines if the fruit fly will develop as a female or a male. The manner in which the sex-lethal mRNA is spliced depends on the signal that is received. Females remove the third exon from the sex-lethal mRNA, whereas males leave the third exon in the mRNA. This is one example of alternative splicing. The female-specific mRNA can be translated by ribosomes to make a fully functional sex-lethal protein. This protein promotes female body development. The male-specific mRNA contains a stop codon in the third exon. This causes the ribosome to stop synthesis of the male's sex-lethal protein earlier than in the female version of the sex-lethal protein. The resulting protein is small and has no function. With no sex-lethal protein activity, the fruit fly develops as a male.
Epigenetics
Epigenetics is the study of changes in gene expression caused by factors other than alterations in a cell’s DNA. The term epigenetics actually means “in addition to genetics,” (i.e., nongenetic factors that cause a cell’s genes to express themselves differently). When an epigenetic change occurs, it might last for the life of the cell and can even be passed on to the next generation. This is what happens when a cell (e.g., stem cell) undergoes the process of differentiation. Stem cells are called pluripotent because they have the potential to be any kind of cell found in the body (i.e., muscle, bone, or skin cell). However, once they become differentiated they lose the ability to become other kinds of cells, and so do the cells they produce by cell division. For example, if a pluripotent cell were to express muscle protein genes and not insulin protein genes, it would differentiate into a muscle cell, not an insulin-producing cell.
Four examples of events that can cause an epigenetic effect are:
1. adding a methyl group to a cytosine in the gene changes it to methylcytosine. Since methylcytosine cannot be read during translation, the gene is turned off.
2. altering the shape of the histones around the gene. Modifying histones ensures that a differentiated cell would stay differentiated, and not convert back into being a pluripotent cell.
3. having the protein that has already been transcribed return to the gene and keep it turned on.
4. splicing RNA into sequences not originally determined by the gene.
Some compounds are considered epigenetic carcinogens (i.e., they are able to cause cells to form tumors), but they do not change the nucleotide sequence of a gene. Examples include certain chlorinated hydrocarbons used as fungicides and some nickel-containing compounds.
8.5. CONCEPT REVIEW
14. Provide two examples of how a cell uses transcription to control gene expression.
15. Provide an example of why it is advantageous for a cell to control gene expression.