CHEMICAL BIOLOGY
Methylerythritol Phosphate Pathway for the Formation of Isoprene Units
Michel Rohmer, Universite Louis Pasteur/CNRS, Institut de Chimie, Strasbourg, France.
doi: 10.1002/9780470048672.wecb631
The carbon skeleton of isoprenoids is derived from the branched C5 skeleton of isoprene. Isopentenyl diphosphate and dimethylallyl diphosphate represent the biologic equivalents of isoprene. From research on cholesterol biosynthesis in liver tissues and on ergosterol in yeast, mevalonate was accepted as the universal precursor of isoprenoids. However this assertion is inaccurate. Incorporation of labeled acetate and glucose isotopomers into bacterial isoprenoids and into diterpenes of ginkgo embryos indicated fortuitously the existence of an alternative mevalonate-independent route. Its full elucidation required experiments using 13C- and 2H-labeled precursors followed by extensive nuclear magnetic resonance analyses as well as a combination of biochemical and molecular biology methods. These additional studies revealed a complete set of novel unsuspected enzymes.
Isoprenoids represent the largest family of natural products, with an exceptional structural diversity. Isoprenoids are present in all living organisms. This group includes essential metabolites, such as sterols 27 (Fig. 6) of the eukaryotic plasma membranes, prenyl chains of the quinones 22 and 23 from electron transport chains, and carotenoids 25 from the photosynthetic apparatus in the plant chloroplasts, or in the phototrophic bacteria. Isoprenoids also include secondary metabolites of a more restricted distribution and with a less obvious physiologic significance. Their carbon skeleton can be derived from the combination of C5 subunits with the branched skeleton of isoprene.
Steps for the Formation of Isoprene Units
The biologic precursors of isoprene units are isopentenyl diphosphate 7 (IPP) and dimethylallyl diphosphate 8 (DMAPP). These precursors can be obtained by two different metabolic pathways: the mevalonate (MVA) pathway (Fig. 1), which was the first one to be elucidated, and the long-overlooked methylerythritol phosphate (MEP) pathway (Fig. 3) (1, 2).
Mevalonate (MVA) pathway
Labeling experiments, performed mainly with liver tissues and yeast to elucidate the biosynthesis of cholesterol and ergosterol (27, Fig. 6), led to the discovery of the mevalonate pathway leading to IPP 7 and DMAPP 8 (Fig. 1). In this pathway, isoprene units are derived, like fatty acids, from acetyl-coenzyme A 1. The key intermediate is mevalonate 4, which results from the reduction of hydroxymethylglutaryl-coenzyme A 3 catalyzed by the HMG-CoA reductase, and represents the committed step of the pathway. Confirmation of this pathway was obtained for the biosynthesis of plant sterols, triterpenes, and sesquiterpenes.
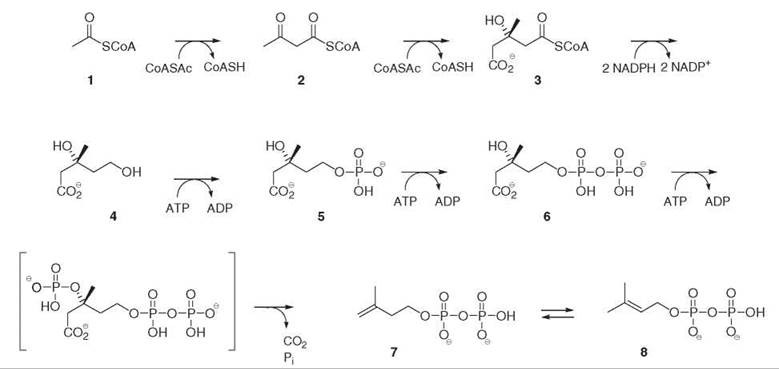
Figure 1. Mevalonate 4 pathway for the biosynthesis of isopentenyl diphosphate 7 and dimethylallyl diphosphate 8.
Discrepancies with the MVA pathway and the discovery of an alternative route: the MEP pathway
The MVA pathway was accepted as the unique biosynthetic pathway for the formation of all isoprenoids in all living organisms. Discrepancies with this general assertion appeared, however, as early as the 1950s (1, 2). For instance, 14C-labeled MVA was not incorporated into chloroplast isoprenoids (e.g., carotenoids 25 and phytol 24 from chlorophylls; Fig. 6), whereas it was well incorporated into phytosterols 27 synthesized in the cytoplasm. Unexpected labeling patterns were found in the prenyl chain of ubiquinone 22 in Escherichia coli at incorporation of 14C-labeled acetate. Finally, the labeling pattern in an isoprene unit from the sesquiterpenic pentalenene 21 series from a Streptomyces species at incorporation of uniformly labeled [U-13C6]glucose was not in accord with what was expected from the MVA pathway.
The discovery of an alternative mevalonate-independent route to isoprene units was a fortuitous, nonprogrammed, and nonprogrammable side product of two independent biosynthetic investigations: on triterpenoids of the hopane series 20 in bacteria (1) and on diterpenes of the ginkgolide 26 and bilobalide series in the higher plant Ginkgo biloba (3). In this pathway (Fig. 2), isoprene units are derived directly from carbohydrate metabolism, with pyruvate 9 and glyceraldehyde 3-phosphate 10 as starting materials and with MEP 12 as the committed intermediate.

Figure 2. Methylerythritol phosphate 12 pathway for the biosynthesis of isopentenyl diphosphate 7 and dimethylallyl diphosphate 8.
Significance of the MEP pathway
The MEP pathway remained overlooked for nearly 40 years. The MEP pathway is present in most eubacteria, including pathogens and opportunistic pathogens, as shown first by biochemical evidence and later by comparison of gene sequences. It is present in all phototrophic organisms, but its presence is restricted to the chloroplasts; The MVA pathway operates in the cytoplasm. It is also found in phylogenetically related nonphotosynthetic taxa, such as the Plasmodium spp., which are the parasites responsible for malaria possessing apicoplasts (plastid-derived organelles).
The MEP pathway revealed a set of seven novel enzymes that correspond to unannotated genes with unprecedented reaction mechanisms, especially the last two enzymes (GcpE and LytB). These two enzymes are characterized by a Fe/S cluster, which catalyzes the conversion of a 1,2-diol derivative into an olefin and the conversion of an allylic alcohol into an olefin.
The MEP Pathway
Several aspects of the MEP pathway reaction sequence are unexpected, including the conversion of two highly oxidized carbohydrate derivatives, pyruvate 9 and glyceraldehyde phosphate 10 (GAP), into two monounsaturated alcohol diphosphates, IPP 7 and DMAPP 8 (Fig. 2).
1-deoxy-D-xylulose 5-phosphate (DXP), the First C5 Intermediate and DXP Synthase
The first step of the MEP pathway is to form DXP 11 by the DXP synthase (DXS) from pyruvate 9 and GAP 10 (2). This reaction could be postulated from the knowledge of the origin of the carbon atoms in isoprene units. Incorporation of deuterium-labeled isotopomers into the prenyl chain of ubiquinone 22 and menaquinone 23 (Fig. 6) from E. coli provided the first evidence for the involvement of deoxyxylulose (DX) in isoprenoid biosynthesis. This observation was extended to plastid isoprenoids in plant systems. Free DX is usually well incorporated into the MEP pathway, with the pentulose being phosphorylated by a nonspecific xylulose kinase in bacteria as well as in plants (4). The DXS gene of this enzyme was identified because of its significant homologies with those of transketolases. Indeed, like the transketolases, DXS uses thiamin diphosphate as a cofactor and catalyzes the condensation of (hydroxyethyl)thiamin diphosphate 17, which results from pyruvate decarboxylation with the carbonyl group of GAP (Fig. 3) (5).
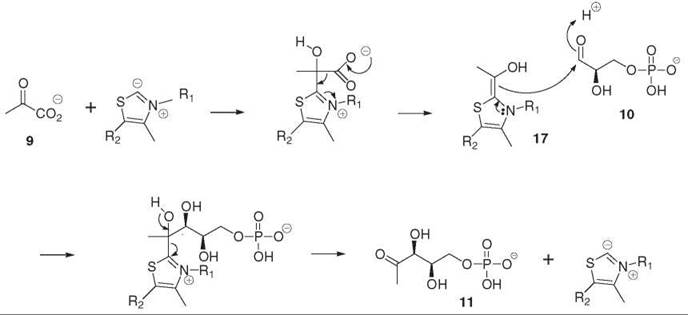
Figure 3. Formation of deoxyxylulose phosphate 11 (DXP) from pyruvate 9 and glyceraldehyde phosphate 10 by the DXP synthase.
DXP reducto-isomerase, the committed step of the MEP pathway and MEP
The incorporation of [4,5-13C2]glucose or [U-13C6]glucose into the hopanoids 20 (Fig. 6) of the bacterium Zymomonas mobilis sheds light on an intramolecular rearrangement step involved in the formation of isoprene units. This rearrangement was later confirmed by the incorporation of DX isotopomers with multiple labeling into the prenyl chains of the quinones 22 and 23 (Fig. 6) in E. coli or phytol 24 from Catharanthus roseus cell cultures (2). This reaction was first written as an acid-catalyzed rearrangement of an α-ketol (Fig. 4a), followed by the NADPH-dependent reduction of the resulting carbonyl derivative. An analogy was made between the corresponding reaction found in the biosynthesis of branched chain amino-acids. A 2-C-methyl-D-erythritol 4-phosphate 12 derivative is the product of this rearrangement. Deuterium-labeled free 2-C-methyl-D-erythritol (ME) was incorporated into the prenyl chain of ubiquinone and menaquinone from wild-type E.coli, but with a much lower yield than DX. This result suggests its correlation with the biosynthesis of isoprene units. In E. coli mutants with a dxs deletion, this deletion was rescued by the addition of synthetic deuterated ME to the culture medium. Incorporation was quantitative, as all isoprene units were derived from the synthetic ME. Free ME added to the culture medium is used only by a few bacteria. In Salmonella enterica, the sorbitol phosphotransferase system is responsible for the phosphorylation and transport of free ME. An analysis of E. coli mutants auxotrophic to ME enables the identification of the DXP reducto-isomerase gene (dxr). As expected, this enzyme catalyzes the rearrangement of DXP into methylerythrose 4-phosphate 18 (Fig. 4) and catalyzes the concomitant NADPH-dependent reduction of the intermediate aldehyde into MEP. The enzyme requires a divalent cation, such as Mg2+ or Mn2+. The postulated aldehyde intermediate 18 has never been observed directly, but synthetic methylerythrose phosphate 18 is converted by DXR into DXP in the presence of NADP+ and into MEP in the presence of NADPH. The addition of NADPH and DXP follows an ordered mechanism with NADPH binding before DXP. The stereochemistry of the reduction step is known. The pro-S hydrogen of NADPH is transferred onto the re face of the aldehyde intermediate 18 (6). 1-Fluoro DXP is a poor substrate of DXR. Although no definitive conclusion could be drawn from this conversion, a careful kinetic analysis of the conversion of this substrate analog suggests an alternative retro-aldol/aldol reaction for the rearrangement step (Fig. 4b) (7) which was confirmed by the secondary isotope effects observed upon incubation of DXP with deuterium labeling at C-3 or C-4 (8).
Fosmidomycin 19 (Fig. 4) and its analogs are strong inhibitors of the MEP pathway, and they inhibit strongly the DXR. According to the X-ray structure of the E. coli DXR, which crystallizes in the presence of fosmidomycin, the antibiotic acts as an analog of the DXP substrate rather than as an analog of the intermediate methylerythrose phosphate (9).

Figure 4. Deoxyxylulose phosphate isomero-reductase (DXR) catalyzed conversion of DXP 11 into MEP 12: (a) α-ketol rearrangement, (b) retro-aldol/aldol reaction. Fosmidomycin 19, a DXR inhibitor.
From methylerythritol phosphate to methylerythritol cyclodiphosphate
No obvious indications were available to identify the next steps after the formation of MEP (2, 10, 11). The incubation of synthetic tritium-labeled MEP with a crude cell-free system from E. coli and with a cocktail of all plausible cofactors and the analysis of the radioactive metabolites suggested the formation of a ME/nucleotide adduct, which could not be characterized fully. A systematic search in gene libraries for a gene encoding an enzyme that uses a polyol phosphate and a nucleotide triphosphate as substrates led to the acsI gene from Haemophilus influenzae encoding an enzyme coupling ribitol 5-phosphate with cytidine 5’-triphosphate (CTP) and yielding the 5’-diphosphocytidyl adduct of ribitol. This gene presented high homologies with the unannotated ygbP gene from E. coli. The corresponding protein was tested in the presence of MEP 12 and nucleotide triphosphates, with the most efficient one being CTP, and catalyzed the formation of the 4-diphosphocytidyl adduct of ME (Fig. 2). In E. coli, the ygbP gene was accompanied by two additional genes, ycbB and ygbB, which together constitute a small cluster. In fact, these two genes coded for the next two enzymes of the pathway. The YchB protein catalyzed the ATP-dependent phosphorylation of the tertiary hydroxyl group of 4-diphosphocytidyl ME 13 yielding 4-diphosphocytidyl ME 2-phosphate 14. The YgbP protein converted the latter intermediate into ME 2,4-cyclodiphosphate 15 with the elimination of cytidine 5’-monophosphate (Fig. 2). Interestingly, in many bacteria, the ygbP and the ygbB genes yield a fusion protein, which corresponds to a bifunctional enzyme catalyzing the two steps, which are performed by two distinct enzymes in E. coli or in plants (12).
From ME Cyclodiphosphate to IPP and DMAPP
In the conversion of MEP 12 into ME cyclodiphosphate 15, the oxidation state of the C5 branched carbon skeleton does not change. The main modification is the introduction of a good leaving group at C-2 on the ME skeleton, which implies that the additional steps include elimination and reduction to afford the unsaturated alcohol diphosphates, IPP and DMAPP, from a tetrol derivative, which is unprecedented in enzyme reactions. Only two additional genes, gcpE and lytB, accompanied all above-mentioned genes of the MEP pathway, making them candidates for the catalysis of the final steps. The amino acid sequence of the two proteins encoded by the gcpE and lytB genes reveals the signature of [Fe4S4] proteins, i.e., with three conserved cysteines involved in the fixation of the cluster in the active sites (2, 13). Most of these proteins are oxygen sensitive, and enzyme tests must be performed under an inert argon atmosphere. GcpE converts ME cyclodiphosphate 15 into 4-hydroxy DMAPP 16, which is in turn converted into IPP 7 and DMAPP 8 in an estimated 5:1 ratio (Fig. 2). The first reaction corresponds with the conversion of a diol derivative into an olefin; the [Fe4S4] cluster in a reduced state acts as an electron donor and probably acts as a Lewis acid for the elimination of the C-3 hydroxy group of ME cyclodiphosphate. The second reaction is the conversion of an allylic alcohol into an olefin. Again the reduced [Fe4S4] cluster of LytB is involved in a reductive one-electron transfer process and acts as a Lewis acid for the elimination of the C-4 hydroxy group of 4-hydroxy DMAPP 16 (Fig. 2).
Both GcpE and LytB must be associated with a reducing system to convert the oxidized [Fe4S4]2+ cluster into the active reduced [Fe4S4]1+ form (Fig. 3), which can be performed with the isolated enzymes from E. coli either by the biologic system flavodoxin/flavodoxin reductase/NADPH, by the semiquinone radical of 5-deazaflavin, or by dithionite. In cyanobacteria, and in plant chloroplasts where flavodoxin is absent, this reducing role can be performed by ferredoxin as shown for the GcpE enzyme, the electron flow coming directly from photosynthesis in the light or from catabolic metabolism in the dark (14).
Elucidation of the MEP Pathway: Key Experiments
The first results from studies the MEP pathway were obtained from in vivo incorporation experiments of 13C labeled glucose isotopomers with bacteria or ginkgo embryos, which determined the origin of the carbon atoms of the isoprene units and which resulted in a reasonable biogenetic scheme. This step was followed by biochemical experiments that were designed to validate the aforementioned scheme and were led to the identification of the first two intermediates, deoxyxylulose phosphate (DXP) and MEP, and the identification of enzymes, deoxyxylulose phosphate synthase (DXS) and DXP reducto-isomerase (DXR). Later steps were identified by using a combination of molecular biology and biochemical methods, including enzyme tests as mentioned in the previous paragraph. Most experiments were cited for the elucidation of the pathway. Three sets of key experiments are described in detail.
Incorporation of 13C-labeled glucose into the hopanoids of Zymomonas mobilis and into the diterpenoids of ginkgo embryos: the origin of carbon atoms in isoprene units
Incorporation studies of 13C-labeled precursors require extensive nuclear magnetic resonance (NMR) measurements. The signals of 13C-NMR-spectra of the analyzed metabolites must be assigned fully. The 13C-NMR spectra of the reference compound of natural abundance (1.1%) and of the labeled metabolite must be recorded in the same conditions. A comparison between the relative signal intensities of the two spectra indicates that the carbon atom is labeled and indicates the magnitude of the isotope enrichment.
The incorporation of 13C-labeled acetate into bacterial hopanoids 20 (Fig. 6) revealed unambiguously a labeling pattern that was not compatible with the MVA pathway. The incorporation of 13C-labeled glucose isotopomers into the isoprene units of the hopanoids from the bacterium Zymomonas mobilis determined the origin of all the carbon atoms of the isoprene units (Fig. 5a) (1, 2). This bacterium only uses glucose as a carbon and energy source via the Entner-Doudoroff pathway. It has no tricarboxylic acid cycle and does not convert pyruvate into glyceraldehyde phosphate. These metabolic peculiarities facilitated a retrobiosynthetic analysis of the labeling pattern, which suggested that the C5 isoprene skeleton was formed from a C2 moiety derived from pyruvate decarboxylation and from a C3 subunit derived from a triose phosphate. The incubation of doubly labeled [4,5-13C2]glucose showed that the coupling of the labeled carbon atoms was preserved in the isoprene units, which indicates that they were introduced together via a single precursor and suggests that an intramolecular rearrangement performed the insertion of the C2 pyruvate-derived subunit between the carbon atoms derived from C-4 and C-5 of glucose. This key experiment excluded the MVA pathway for the formation of the bacterial isoprene units and was the signature of an alternative biosynthetic route.
Similar experiments performed on ginkgo embryos also revealed an unexpected labeling pattern of the isoprene units of the diterpenoid skeletons. In this case, glucose is metabolized via glycolysis and a retrobiosynthetic analysis was in accordance with the formation of isoprene units from pyruvate and from a triose phosphate derivative (Fig. 5b) (3).
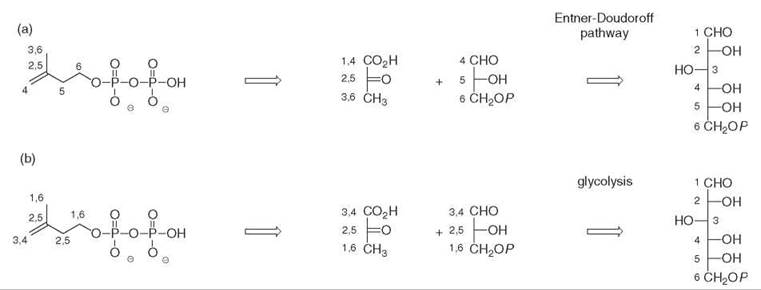
Figure 5. Origin of carbon atoms from glucose in the isoprene units of (a) the hopanoids of Zymomonas mobilis, (b) the diterpenoids of Ginkgo biloba.

Figure 6. Isoprenoids from bacteria (bacteriohopanepolyols, 20; pentalenolactone, 21; ubiquinone, 22; menaquinone, 23) and from higher plants (phytol, 24; β-carotene, 25; ginkgolide, 26; sterols, 27).
Incorporation of 2H-labeled deoxyxylulose and methylerythritol into terpenoids from bacteria and from plant plastids
The incorporation of 13C-labeled glucose isotopomers showed that a pyruvate and a triose phosphate derivative were the precursors of isoprene units in the alternative route. The triose phosphate derivative was identified as glyceraldehyde phosphate using E. coli mutants; each lacks a single enzyme of the triose phosphate metabolism (allowing the interconversion of pyruvate and glycerol) and grows either in the presence of no-labeled pyruvate and 13C-labeled glycerol or in the presence of a 13C-labeled pyruvate and nonlabeled glycerol. These initial results led to the identification of the first two C5 intermediates of the pathway.
1-Deoxy-D-xylulose is a known natural product. It was first isolated from the fermentation broth of a Streptomyces and later shown to be a precursor of pyridoxol. Its structure can be deduced biogenetically from pyruvate and glyceraldehyde phosphate (Figs. 2 and 3). Two 2H-labeled DX isotopomers were synthesized chemically. They were incorporated efficiently into the prenyl chains of ubiquinone and menaquinone by wild-type E. coli, indicating that a DX derivative is an isoprenoid precursor (2, 3).
Free methylerythritol is a widespread polyol in plants. The formation of the isoprene skeleton via the alternative route involves an intramolecular rearrangement. The branched carbon skeleton of ME can be deduced from the rearrangement of the straight chain DX (Fig. 4). Deuterium-labeled ME isotopomers were synthesized chemically and were incorporated into the prenyl chains of the E. coli quinones (2).
E. coli constructs using MVA for the identification of MEP pathway genes
Many isoprenoids are essential metabolites in living cells. Inhibition of their biosynthesis is lethal, which implies that any deletion or any major mutation of a gene of the MEP pathway will be lethal and must be rescued by another source of precursor for IPP and DMAPP. In E. coli, this can be performed by introducing the genes of the MVA pathway, which is absent in this bacterium (15).
For instance, the deletion of the dxs gene, which encodes the enzyme catalyzing the conversion of DXP 11 into MEP 12, is lethal. This deletion can be rescued two ways: by adding synthetic ME, which is phosphorylated in vivo and enters the MEP pathway, or by introducing the genes of enzymes which allow the conversion of MVA into IPP. For this reason, three genes must be introduced in the construct, encoding, respectively, mevalonate 4 kinase, phosphomevalonate 5 kinase, and diphosphomevalonate 6 decarboxylase (Fig. 1).
This approach has been used to check that gcpE and lytB are involved in the MEP pathway. These genes accompanied regularly the other known genes of the pathway in the completely sequenced bacterial genomes. The deletion of either of these genes was lethal, which indicates that they are essential but was rescued by the insertion of the three above-mentioned genes of the MVA pathway, indicating that they are involved in the biosynthesis of IPP and DMAPP (2).
Future Developments
Inhibition of the MEP pathway: toward novel antibacterial and antiparasitic drugs
The MEP pathway is the only pathway involved in the biosynthesis of essential isoprenoids in pathogenic bacteria and in parasites, and it is absent in animals and in humans. Therefore, any enzyme of this pathway is a potential target for a novel type of antimicrobial drugs (16, 17). This concept has been validated by the mode of action of fosmidomycin, a natural antibiotic that inhibits the second step of the MEP pathway catalyzed by the deoxyxylulose phosphate reducto-isomerase (DXR).
Overexpression of the MEP pathway: toward improved production of plant terpenoids of economic value
The MEP pathway is the starting material of major plant terpenoids of economic value: e.g., monoterpenes from essential oils, diterpenoids with potent biologic activity (taxol, ginkgolides), vitamins (tocopherol), or vitamin precursors (carotenoids). The first attempts to overexpress some of its genes (especially dxs or dxr) led to enhanced carotenoid production in tomato (18) or monoterpenes in mint (19).
Cross-talk between the MVA and the MEP pathway: A Novel Aspect for the Regulation of Terpenoid Biosynthesis in Plants
Finally, the dichotomy between the cytoplasmic mevalonate pathway and the plastidial MEP pathway is not strict. An exchange of metabolites occurs between the two compartments at the level of C5, C10, and C15 prenyl diphosphates. The inhibition of one pathway (e.g., by mevinolin for the MVA pathway or by fosmidomycin for the MEP pathway) can be complemented by the other route either partially, as in most tested plant systems, or even completely, in the case of the tobacco Bright Yellow-2 cell cultures (20). In addition, a regulation of the two pathways by the nycthemeral cycle is suggested; the MEP pathway is activated in the presence of light and is activated by photosynthesis (21). Cross-talk and light regulation lead to new findings in the regulation of isoprenoid biosynthesis in plants.
References
1. Rohmer M. A mevalonate-independent route to isopentenyl diphosphate. In: Comprehensive Natural Product Chemistry. Isoprenoids Including Carotenoids. Cane, David E. ed. 1999. Pergamon, Oxford, UK.
2. Rohmer M. Diversity in isoprene unit biosynthesis: the methylerythritol phosphate pathway in bacteria and plastids. Pure Appl. Chem. 2007; 79:739-751.
3. Schwarz M, Arigoni D. Ginkgolide biosynthesis. In: Comprehensive Natural Product Chemistry. Isoprenoids Including Carotenoids. Cane David E. ed. 1999. Pergamon, Oxford, UK.
4. Hemmerlin A, Tritsch D, Hartmann MA, Pacaud K, Hoeffler JF, Van Dorsselaer A, Rohmer M, Bach TJ. A cytosolic Arabidopsis thaliana D-xylulokinase catalyzes the phosphorylation of 1-deoxy-D-xylulose into a precursor of the plastidial isoprenoid pathway. Plant Physiol. 2006; 142:441-457.
5. Eubanks LM, Poulter CD. Rhodobacter capsulatus 1-deoxy-D-xylulose 5-phosphate synthase: steady-state kinetics and substrate binding. Biochemistry 2003; 42:1140-1149.
6. Proteau PJ. 1-Deoxy-D-xylulose 5-phosphate reductoisomerase: an overview. Bioorg. Chem. 2004; 32:483-493.
7. Fox DT, Poulter CD. Mechanistic studies with 2-C-methyl-D-erythritol 4-phosphate synthase from Escherichia coli. Biochemistry 2005; 44:3860-3868.
8. Wong U, Cox RJ. The chemical mechanism of D-1-deoxyxylulose-5-phosphate reducto-isomerase from Escherichia coli. Angew. Chem. Int. Ed. 2007; 46:4926-4929.
9. Steinbacher S, Kaiser J, Eisenreich W, Huber R, Bacher A, Rohdich F. Structural basis of fosmidomycin action revealed by the complex with 2-C -methyl-D-erythritol 4-phosphate synthase (IspC). J. Biol. Chem. 2003; 278:18401-18407.
10. Rohdich F, Kis K, Bacher A, Eisenreich W. The non-mevalonate pathway of isoprenoids: genes, enzymes and intermediates. Curr. Opin. Chem. Biol. 2001; 5:535-540.
11. Eisenreich W, Bacher A, Arigoni D, Rohdich F. Biosynthesis of isoprenoids via the non-mevalonate pathway. Cell. Mol. Life Sci. 2004; 61:1401-1426.
12. Gabrielsen M, Rohdich F, Eisenreich W, Grawert T, Hecht S, Bacher A, Hunter WW. Biosynthesis of isoprenoids. A bifunctional IspDF enzyme from Campylobacter jejuni. Eur. J. Biochem. 2004; 271:3028-3025.
13. Seemann M, Rohmer M. Isoprenoid biosynthesis via the methyl-erythritol phosphate pathway: GcpE and LytB, two novel iron/sulphur proteins. C.R. Chimie 2007. In press.
14. Seemann M, Tse Sum Bui B, Wolff M, Miginiac-Maslow M; Rohmer M. isoprenoid biosynthesis in plant chloroplasts via the MEP pathway: direct thylakoid/ferredoxin-dependent photoreduction of GcpE/IspG. FEBS Lett. 2006; 580:1547-1552.
15. Rodriguez-Concepcion M, Campos N, Lois LM, Maldonaldo C, Hoeffler JF, Grosdemange-Billiard C, Rohmer M, Boronat A. Genetic evidence of branching in the isoprenoid pathway for the production of isopentenyl diphosphate and dimethylallyl diphosphate. FEBS Lett. 2000; 433:328-332.
16. Jomaa H, Wiesner J, Sanderbrand S, Altincicek B, Weidemeyer C, Hintz M, Tiirbachova I, Eberl M, Zeidler J, Lichtenthaler HK, Soldati D, Beck E. Inhibitors of the nonmevalonate pathway of isoprenoid biosynthesis as antimalarial drugs. Science 1999; 285:1573-1576.
17. Rohmer M, Grosdemange-Billiard C, Seemann M, Tritsch D. Isoprenoid biosynthesis as a novel target for antibacterial and antiparasitic drugs. Curr. Opinion Invest. Drugs 2004; 5:154-162.
18. Enfissi EMA, Fraser PD, Lois LM, Boronat A, Schuch W, Bramley PM. Metabolic engineering of the mevalonate and non-mevalonate isopentenyl diphosphate-forming pathways for the production of health-promoting isoprenoids in tomato. Plant Biotechnol. J. 2003; 3:17-27.
19. Mahmoud SS, Croteau RB. Metabolic engineering of essential oil yield and composition in mint by altering expression of deoxyxylulose phosphate reductoisomerase and menthofuran synthase. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:8915-8920.
20. Hemmerlin A, Hoeffler JF, Meyer O, Tritsch D, Kagan IA, Grosdemange-Billiard C, Rohmer M, Bach TJ. Cross-talk between cytosolic mevalonate and plastidial methylerythritol phosphate pathways in tobacco Bright Yellow-2 cells. J. Biol. Chem. 2003; 278:26666-26676.
21. Dudareva N, Andersson S, Orlova I, Gatto N, Reichelt M, Rhodes D, Boland W, Gershenzon J. The nonmevalonate pathway supports both monoterpene and sequiterpene formation in snapdragon flowers. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:933-938.
Further Reading
Bacher A, Rieder C, Eichinger D, Arigoni D, Fuchs G, Eisenreich W. Elucidation of novel biosynthetic pathways and metabolic flux patterns by retrobiosynthetic NMR analysis. FEMS Microbiol. Rev. 1999; 22:567-598.
Hemmerlin A, Gerber E, Feldtrauer JF, Wentzinger L, Hartmann MA, Tritsch D, Hoeffler JF, Rohmer M, Bach TJ. A review of tobacco BY-2 cells as an excellent system to study the synthesis and function of sterols and other isoprenoids. Lipids 2004; 39:723-735.
Jux A, Gleixner G, Boland W. Classification of terpenoids according to the methylerythritol phosphate or the mevalonate pathway with natural 12C/13C ratios: dynamic allocation of resources in induced plants. Angew. Chem. Int. Ed. 2001; 40:2091-2093.
Kim D, Filtz MR, Proteau PJ. The methylerythritol phosphate pathway contributes to carotenoid but not phytol biosynthesis in Euglena gracilis. J. Nat. Prod. 2004; 67:1067-1069.
Kuzuyama T,Shimizu T, Takahashi S, Seto H. Fosmidomycin, a specific inhibitor of 1-deoxy-D-xylulose 5-phosphate reductoisomerase in the nonmevalonate pathway for terpenoid biosynthesis. Tetrahedron Lett. 1998; 39:7913-7916.
Lell B, Ruangweerayut R, Wiesner J, Anoumou-Missinou M, Schindler A, Baranek T, Hintz M, Hutchinson D, Jomaa H, Kremsner PG. Fosmidomycin, a novel chemotherapeutic agent for malaria. Antimicrob. Agents Chemother. 2003; 47:735-738.
Lherbet C, Pojer F, Richard SB, Noel JP, Poulter CD. Absence of substrate channeling between active sites in the Agrobacterium tume-faciens IspDF and IspE enzymes of the methylerythritol phosphate pathway. Biochemistry 2006; 45:3548-3553.
Rohdich F, Wungsintaweekul J, Fellermeier M, Sagner M, Herz S, Kis K, Eisenreich W, Bacher A, Zenk MH. Cytidine 5-triphosphate-dependent biosynthesis of isoprenoids: YgbP protein of Escherichia coli catalyzes the formation of 4-diphosphocytidyl-2-C-methyl-D-erythritol. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:11758-11763.
Rohmer M, Knani M, Simonin P, Sutter B, Sahm H. Isoprenoid biosynthesis in bacteria: a novel pathway for the early steps leading to isopentenyl diphosphate. Biochem. J. 1993; 295:517-524.
Rohmer M, Seemann M, Horbach S, Bringer-Meyer S, Sahm H. Glyceraldehyde 3-phosphate and pyruvate as precursors of isoprenic units in an alternative non-mevalonate pathway for terpenoid biosynthesis. J. Am. Chem. Soc. 1996; 118:2564-2566.
Schuhr C, Radykewicz Sagner S, Latzel C, Zenk MH, Arigoni D, Bacher A, Rohdich F, Eisenreich W. Quantitative assessment of crosstalk between the two isoprenoid biosynthesis pathways by NMR spectroscopy. Phytochem. Rev. 2003; 2:3-16.
Takahashi S, Kuzuyama T, Watanabe Seto H. A 1-deoxy-D-xylulose reductoisomerase catalyzing the formation of 2-C-methyl- D-erythritol 4-phosphate in an alternative nonmevalonate pathway for terpenoid biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:9879- 9884.
See Also
Antibacterial Drugs, Design of
Iron-Sulfur World
Isoprenoids
Steroid and Triterpene Biosynthesis
Terpenes, Biosynthesis of Terpenoids in Plants