MCAT Biology and Biochemistry: New for MCAT 2015 (2014)
Chapter 7. Eukaryotic Cells
The first cells were prokaryotes. They consisted of a cell membrane and a cell wall surrounding the cytoplasm or cell fluid. All the structures necessary for survival and reproduction floated in the cytoplasm, including the double-stranded circular DNA genome, ribosomes, the enzymes of aerobic and anaerobic metabolism, etc. As evolution proceeded, cell complexity increased. The greatest landmark in the evolution of the cell was the development of membrane-bound compartments within the cytoplasm known as organelles. These served to organize the cytoplasm, with each membrane acting to seal its compartment. The most important organelle is the control center of the cell: the nucleus. In fact, “eukaryotic” is from the Greek “karyon,” meaning “kernel” or “nucleus,” plus the prefix “eu,” meaning “true.” “Prokaryotic” means “before the nucleus” and also implies “before organelles.” All true living organisms are either prokaryotes or eukaryotes. There are three well-defined kingdoms within Domain Eukarya (Plantae, Animalia, and Fungi), and one group of organisms for whom the kingdom classifications are under debate (single- celled eukaryotes … the Protists).
7.1 INTRODUCTION
It would be impossible to understand medicine without sound knowledge of the eukaryotic cell. In this chapter we will examine each of the principal organelles, beginning with the nucleus. Next we will focus on the plasma membrane, then the cytoskeleton, and finally we will finish with a discussion of the cell cycle. You should be able to explain the function of each item labeled in Figure 1 below. Our discussion will be based on the animal cell. Fungi have already been discussed in the previous chapter, and neither plants nor protists (motile unicellular eukaryotes) are covered on the MCAT.
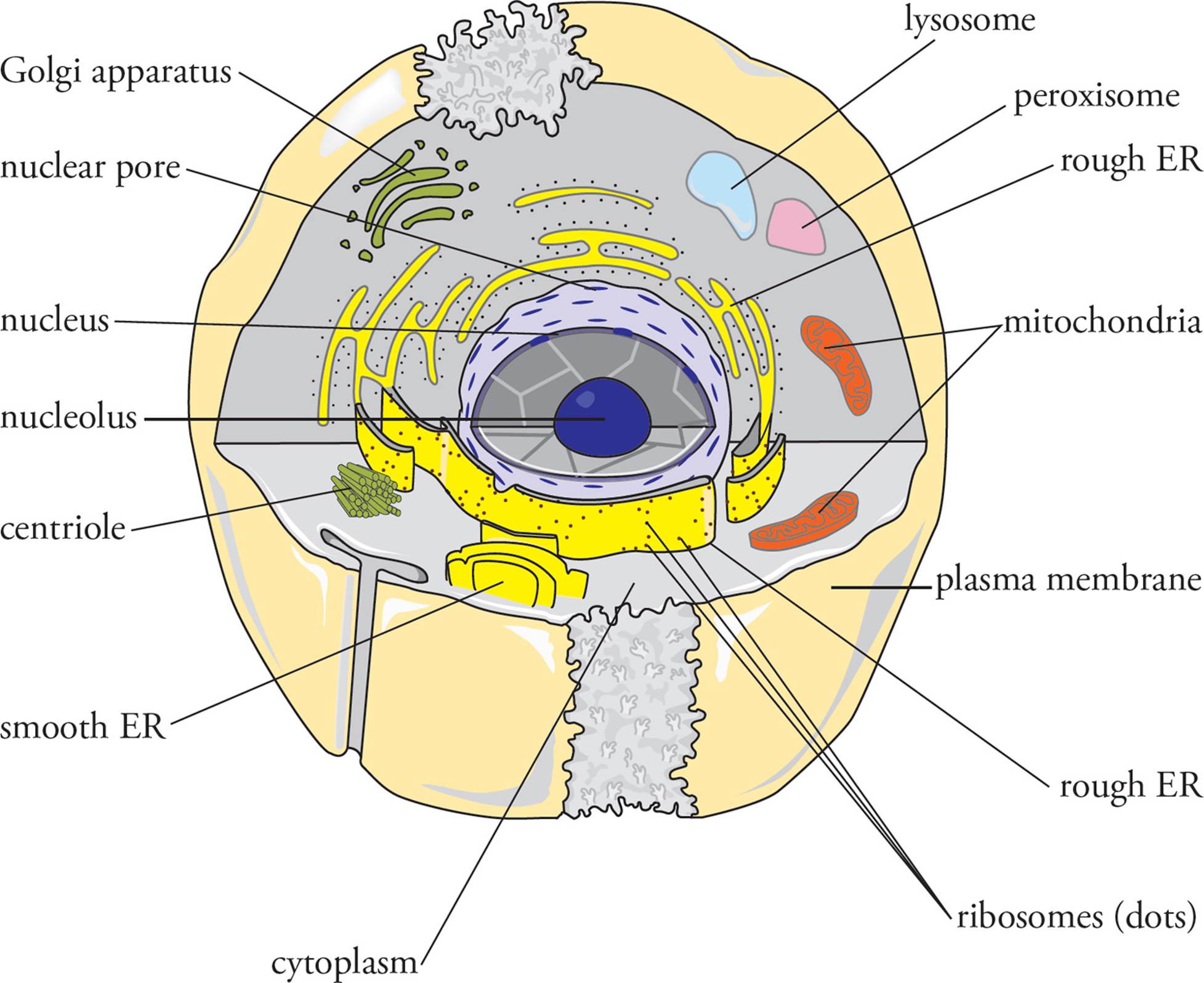
Figure 1 The Eukaryotic Cell
7.2 THE ORGANELLES
An organelle is a small structure within a cell that carries out specific cellular functions. Most organelles are bounded by their own lipid bilayer membrane. The membrane acts like a plastic bag to seal off the contents of the organelle from the rest of the cytoplasm and control what enters and exits. A summary of the major animal cell organelles is given in the Table below:
|
Organelle |
Function (number of membranes surrounding) |
|
nucleus |
contain & protect DNA, transcription, partial assembly of ribosomes (2) |
|
mitochondria |
produce ATP via the Krebs cycle and oxidative phosphorylation (2) |
|
ribosomes |
synthesize proteins (0) |
|
RER |
location of synthesis/modification of secretory, membrane-bound, & organelle proteins (1) |
|
SER |
detoxification & glycogen breakdown in liver; steroid synthesis in gonads (1) |
|
Golgi apparatus |
modification & sorting of protein, some synthesis (1) |
|
lysosomes |
contain acid hydrolases which digest various substances (1) |
|
peroxisomes |
metabolize lipids & toxins using H2O2 (1) |
Table 1 Animal Cell Organelles
The Nucleus
One of the primary features of eukaryotic cells distinguishing them from prokaryotic cells is the nucleus. The nucleus contains the genome surrounded by the nuclear envelope that separates the contents of the nucleus into a distinct compartment, isolated from other organelles and from the cytoplasm. In prokaryotes the genome may be localized in the cell, but without a nuclear envelope to form a separate compartment, the genome remains accessible to the cytoplasm. In prokaryotes, replication, transcription and translation, and everything else all happens in the same compartment (the cytoplasm). In eukaryotes, replication, transcription, and splicing occur in the nucleus, while translation occurs in the cytoplasm.
• If an enzyme that degrades mRNA is injected into the cytoplasm of a cell and all translation ceases, is the cell prokaryotic or eukaryotic?1
• When an enzyme that degrades DNA (DNase) is incubated with intact DNA isolated from an organism, the DNA is degraded. But when DNase is injected into the cytoplasm of cells from the same organism, no effect on the genome is observed. Which one of the following is the best explanation for this?2
A) The cell is a prokaryote; therefore, the genome is inaccessible to cytoplasmic enzymes.
B) The cell is a prokaryote; therefore, the circular genome is resistant to DNase.
C) The cell is a eukaryote; therefore, the genome is inaccessible to cytoplasmic enzymes.
D) The cell is a eukaryote; therefore, the linear genome is resistant to DNase.
The Genome
The eukaryotic genome was introduced in Chapters 5 and 6, but we will touch on it again here, as well as in Chapter 8. Eukaryotic genomes are organized into linear molecules of double-stranded DNA, while the genome of prokaryotes is a single circular DNA molecule. The large size of the typical eukaryotic genome appears to make it necessary to split the genome into pieces, each a separate linear DNA molecule, termed a chromosome. Yeast have 4 different chromosomes, while there are 23 different human chromosomes. Since humans and most adult animals are diploid, they have two copies of each chromosome (except for the sex chromosomes; see Chapter 8). Chromosomes have a centromere near the middle to ensure that newly replicated chromosomes are sorted properly during cell division, one copy to each daughter cell (mitosis and meiosis, this chapter and Chapter 8). Each eukaryotic chromosome also has special structures at both ends termed telomeres. Telomeres have large numbers of repeats of a specific DNA sequence and, with the help of a special DNA polymerase termed telomerase, maintain the ends of the linear chromosomes during DNA replication. [What special problems are there in replicating the 5 ends of linear DNA chromosomes?3]
Within each chromosome is also a portion of the many thousands of genes in the genome as a whole. Genes can be mapped genetically and physically to the chromosome they reside on and to a specific location on that chromosome, a locus. The expression of eukaryotic genes is regulated by specific promoter and enhancer elements of that gene, but can also be affected by the position of the gene on the chromosome. Some regions of a chromosome are folded into densely packed chromatin, termed heterochromatin, within which genes tend to be inaccessible and turned off. Other regions known as euchromatin are more loosely packed (although still packaged into chromatin) and allow genes to be activated (see Chapter 5). [If a retrovirus inserts its genome into regions of heterochromatin and nowhere else, how is this likely to affect the infection process?4]
Finally, the nucleus is not a loose membrane bag with DNA floating inside. If nuclei are treated with DNase and with detergent, an insoluble mesh of protein, known as the nuclear matrix or nuclear scaffold, is left behind. The role of the nuclear matrix may be in part analogous to the role of the cytoskeleton in the cytoplasm: to support and provide overall structure. The matrix may also play a role in regulating gene expression. The DNA in chromosomes is attached to the matrix at specific sites, and these (in some cases) appear to be involved in regulating gene expression or in limiting the effects of promoters and enhancers to discrete chromosomal regions known as domains. The role of the nuclear matrix is an area of ongoing research.
The Nucleolus
The nucleolus (“little nucleus”) is a region within the nucleus which functions as a ribosome factory. There is no membrane separating the nucleolus from the rest of the nucleus. It consists of loops of DNA, RNA polymerases, rRNA, and the protein components of the ribosome. [Would you expect the nucleolus to be larger in cells that are actively synthesizing protein, or in quiescent cells?5 What role would the loops of DNA in the nucleolus play?6]
The nucleolus is the site of transcription of rRNA by RNA pol I. Transcription of mRNA and tRNA is performed by other polymerases in other areas of the nucleus. [Does a similar “division of labor” exist in the prokaryotic cell?7] The ribosome is partially assembled while still in the nucleolus. The protein components of the ribosome are not produced in the nucleolus; they are transported into the nucleus from the cytoplasm (remember that all translation takes place in the cytoplasm). After partial assembly, the ribosome is exported from the nucleus, remaining inactive until assembly is completed in the cytoplasm. This may serve to prevent translation of hnRNA.
The Nuclear Envelope
Surrounding the nucleus and separating it from the cytoplasm is the nuclear envelope, composed of two lipid bilayer membranes. The inner nuclear membrane is the surface of the envelope facing the nuclear interior, and the outer nuclear membrane faces the cytoplasm. The membrane of the endoplasmic reticulum is at points continuous with the outer nuclear membrane, making the interior of the ER (the lumen of the ER) contiguous with the space between the two nuclear membranes. [Is the space between the inner and outer membranes contiguous with the cytoplasm?8]
The nuclear envelope is punctuated with large nuclear pores that allow the passage of material into and out of the nucleus (see Figures 2 and 3). Molecules that are smaller than 60 kilodaltons, including small proteins, can freely diffuse from the cytoplasm into the nucleus through the nuclear pores. Larger proteins cannot pass freely through nuclear pores and are excluded from the nuclear interior unless they contain a sequence of basic amino acids called a nuclear localization sequence. Proteins with a nuclear localization sequence are translated on cytoplasmic ribosomes and then imported into the nucleus by specific transport mechanisms. It also appears likely that RNA is transported out of the nucleus by a specific transport system rather than freely diffusing into the cytoplasm. [If a 15 kD protein has a nuclear localization sequence that is then deleted from its gene, will the mutated protein still be found in the nucleus?9]
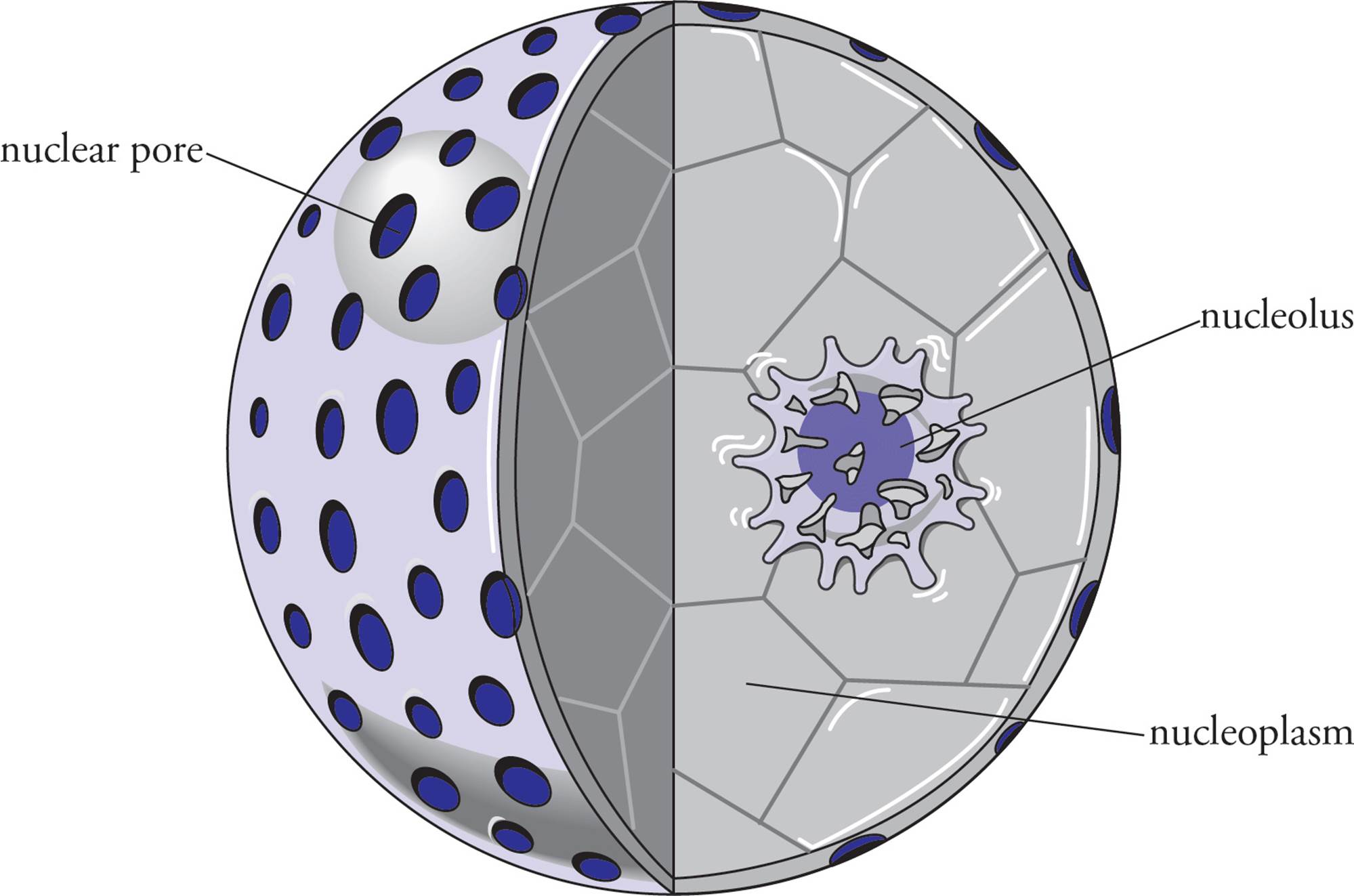
Figure 2 The Nucleus, Showing Pores
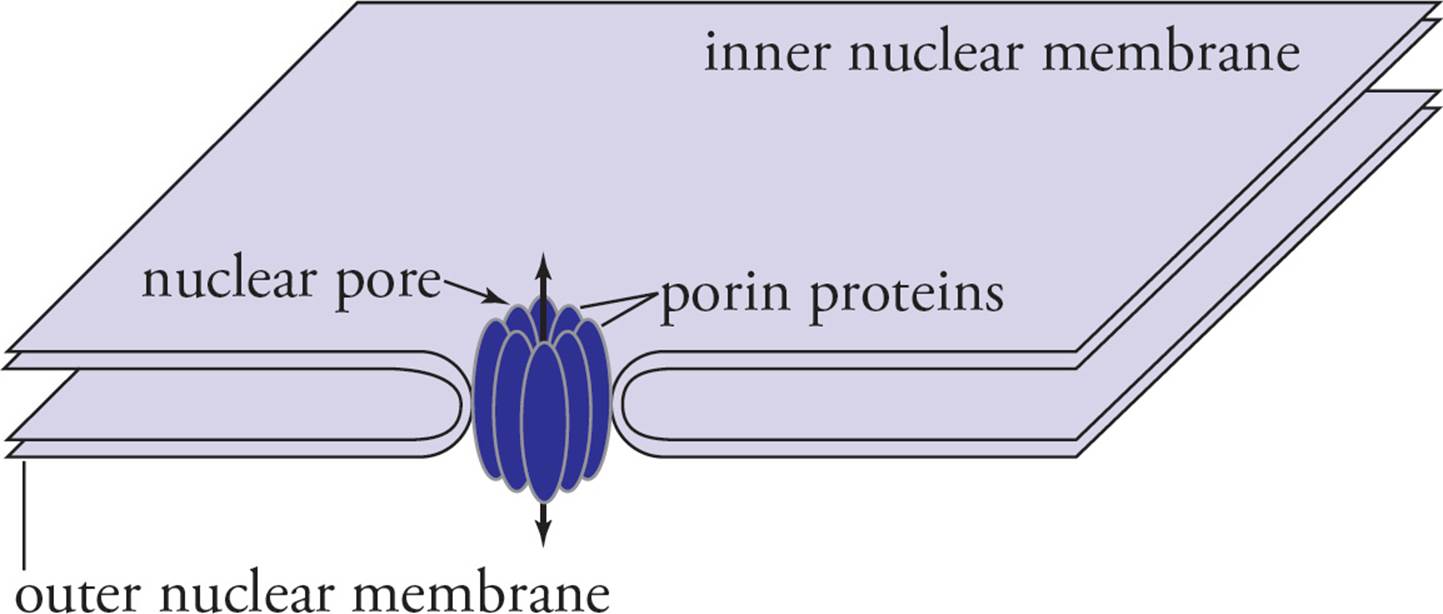
Figure 3 A Nuclear Pore Close-Up
• Which one of the following proteins would NOT be found within the nucleus?10
A) A protein component of the large ribosomal subunit
B) A factor required for splicing
C) A histone
D) An aminoacyl tRNA synthetase
• A researcher injects tiny gold beads into a cell and waits an hour. Then she examines the cell and finds gold beads in the cytoplasm and in the nucleus. When she injects larger gold beads, they are not found in the nucleus. However, when she binds the larger beads to a nuclear localization sequence, she finds that they end up in the nucleus. One can conclude that:11
A) the nuclear localization sequence is lysine-rich.
B) gold beads have an inherent import signal.
C) the nuclear localization mechanism is nonspecific enough to confer nuclear import on gold beads.
D) nuclear import relies primarily on simple diffusion.
Mitochondria
Mitochondria are the site of oxidative phosphorylation (discussed in more detail in Chapter 4). The interior of mitochondria, the matrix, is bounded by the inner and outer mitochondrial membranes (see Figure 4). The matrix contains pyruvate dehydrogenase and the enzymes of the Krebs cycle. The inner membrane is the location of the electron transport chain and ATP synthase and is the site of the proton gradient used to drive ATP synthesis by ATP synthase. The inner membrane is impermeable to the free diffusion of polar substances, like protons, and is folded into the matrix in projections called cristae. The outer membrane is smooth and contains large pores that allow free passage of small molecules. The space between the membranes is called the intermembrane space. ATP produced within mitochondria is transported out into the cytoplasm to drive a great variety of cellular processes. [Why is the inner membrane folded into cristae?12 Are the enzymes of glycolysis found in the matrix?13 If the inner membrane is impermeable, how does pyruvate get into the matrix where pyruvate dehydrogenase is located?14]
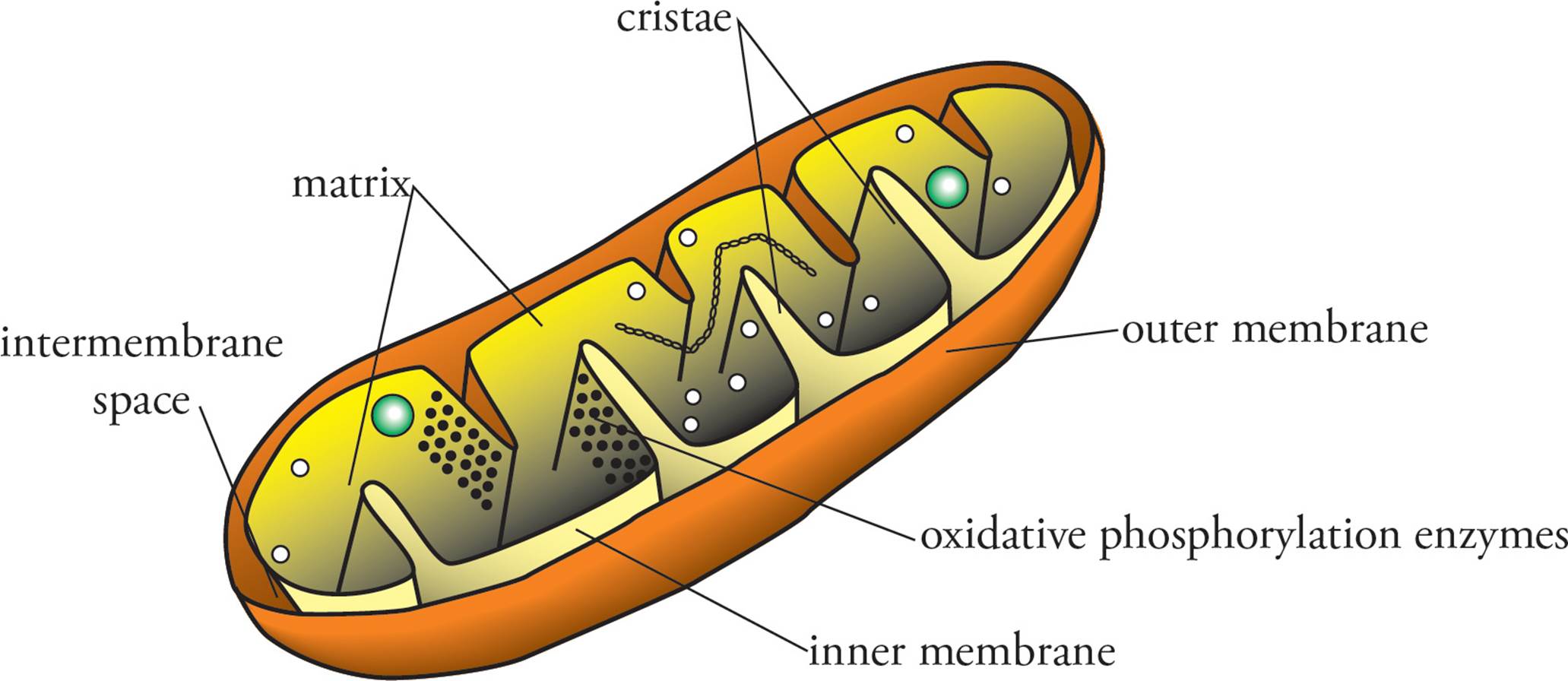
Figure 4 The Mitochondrion
Mitochondria possess their own genome which is far smaller than the cellular genome and consists of a single circular DNA molecule. (Sound familiar?) It encodes rRNA, tRNA, and several proteins, including some components of the electron transport chain and parts of the ATP synthase complex although most mitochondrial proteins are encoded by nuclear genes. Even more curious, mitochondria use a different system of transcription and translation than nuclear genes do. This includes a unique genetic code and unique RNA polymerases, DNA replication machinery, ribosomes, and aminoacyl-tRNA synthetases. In order to explain the fact that mitochondria possess a second system of inheritance, investigators have postulated that mitochondria originated as independent unicellular organisms living within larger cells. This is known as the endosymbiotic theory of mitochondrial evolution (endo = within; symbiotic = living together). In fact, if you compare a mitochondrion to a Gram-negative bacterium, you’ll note that they look pretty similar. Pay attention to where the enzymes of electron transport are located and the genome shape.15 Because many unique mitochondrial polypeptides are encoded by the cellular genome and not the mitochondrial genome, it has been suggested that the genes coding for these proteins may have been transferred to the nuclear genome over time. [What difficulty may be encountered in translation of a mitochondrial gene moved to the nucleus?16]
Mitochondria exhibit maternal inheritance. This means that mitochondria are inherited only from the mother, since the cytoplasm of the egg becomes the cytoplasm of the zygote. (The sperm contributes only genomic [nuclear] DNA.) Maternal inheritance departs from the rules of Mendelian genetics, which state that traits are inherited from both parents (Chapter 8). If a woman has a disease caused by an abnormality in her mitochondrial genome, what are the chances that her children will have the disease (assuming her mate does not have the disease)?17
Endoplasmic Reticulum (ER)
The endoplasmic reticulum (ER) is a large system of folded membrane accounting for over half of the membrane of some cells. There are two types of ER (see Figure 5): rough ER and smooth ER, each with distinct functions. The rough ER is called rough due to the large number of ribosomes bound to its surface; it is the site of protein synthesis for proteins targeted to enter the secretory pathway. The smooth ER is not actively involved in protein processing but can contain enzymes involved in steroid hormone biosynthesis (gonads) or in the degradation of environmental toxins (liver). The membrane of the endoplasmic reticulum is joined with the outer nuclear membrane in places, meaning that the space within the nuclear membranes is continuous with the interior of the ER (the ER lumen). The rough ER plays a key role directing protein traffic to different parts of the cell.
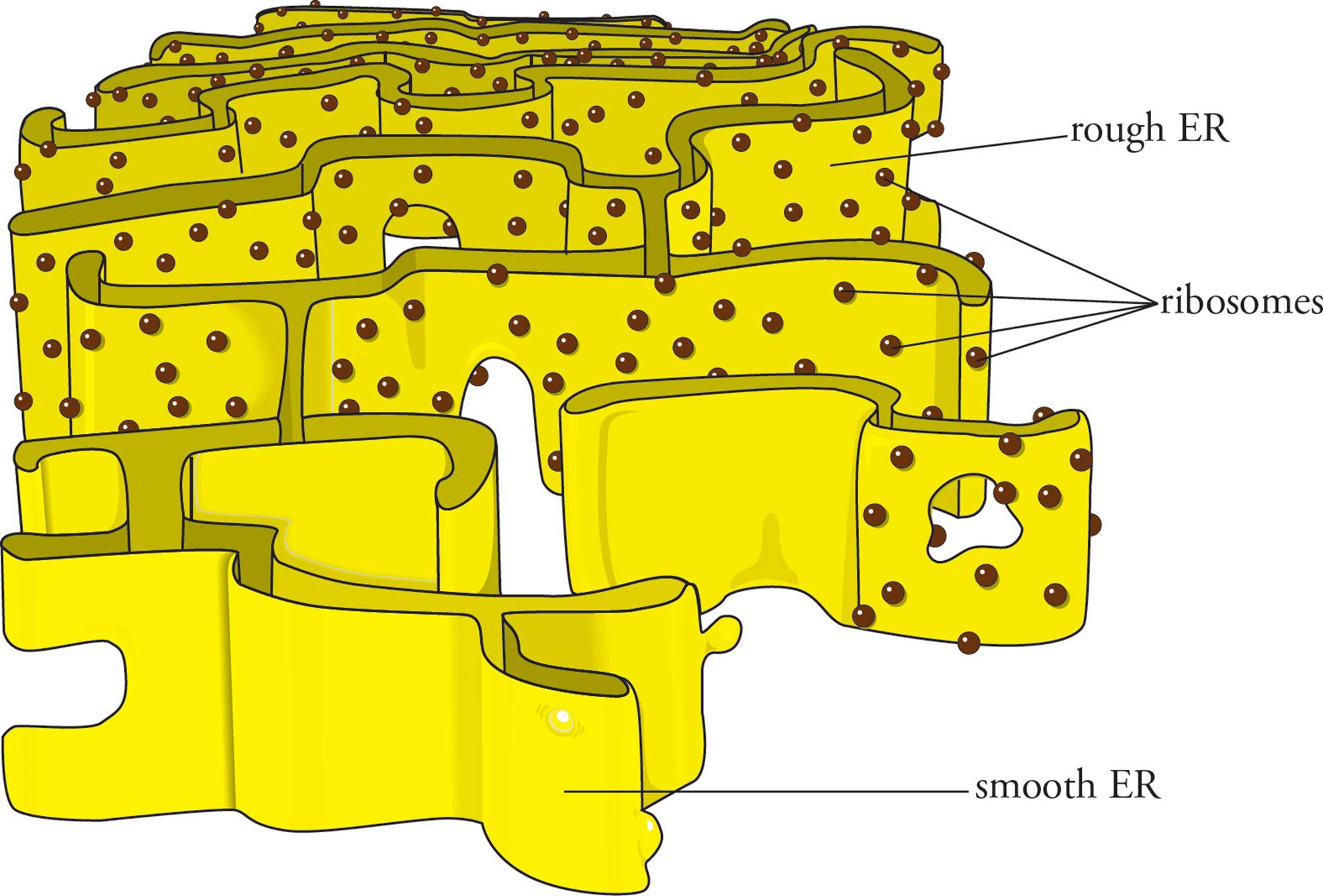
Figure 5 The ER
The Rough ER and the Secretory Pathway
There are two sites of protein synthesis in the eukaryotic cell: either on ribosomes free in the cytoplasm or on ribosomes bound to the surface of the rough ER. Proteins translated on free cytoplasmic ribosomes are headed toward peroxisomes, mitochondria, the nucleus, or will remain in the cytoplasm. Proteins synthesized on the rough ER will end up either 1) secreted into the extracellular environment, 2) as integral plasma membrane proteins, or 3) in the membrane or interior of the ER, Golgi apparatus, or lysosomes. Membrane-bound vesicles pass between these cellular compartments. Since the membranes of these organelles communicate through the traffic of vesicles, the interior of the ER, the Golgi apparatus, lysosomes, and the extracellular environment are in a sense contiguous. Proteins synthesized on the rough ER are transported in vesicles that bud from the ER to the Golgi apparatus, then to the plasma membrane or lysosome. A secreted protein that enters the ER lumen is separated by a membrane from the cytoplasm until the protein leaves the cell.
Whether a protein is translated on the rough ER is determined by the sequence of the protein itself. All proteins start translation in the cytoplasm; however, some proteins (secreted proteins and lysosomal proteins) have an amino acid sequence at their N-terminus called a signal sequence. The signal sequence of a nascent polypeptide is recognized by the signal recognition particle (SRP), which binds to the ribosome. The rough ER has SRP receptors that dock the ribosome-SRP complex on the cytoplasmic surface (along with the nascent polypeptide and mRNA). Translation then pushes the polypeptide, signal peptide first, into the ER lumen. After translation is complete, the signal peptide is removed from the polypeptide by a signal peptidase in the ER lumen. For secreted proteins, once the signal sequence is removed, the protein is transported in the interior of vesicles through the Golgi apparatus to the plasma membrane, where it is released by exocytosis into the extracellular environment.
• The mRNA for a secreted protein encodes a longer protein than is actually observed in the cellular exterior. Why?18
A) The protein was cleaved by a cytoplasmic protease.
B) The mRNA was not spliced properly.
C) The gene encoding the protein contained a nonsense mutation.
D) The signal sequence of the protein was removed in the rough ER.
Integral membrane proteins are processed slightly differently. Integral membrane proteins have sections of hydrophobic amino acid residues called transmembrane domains that pass through lipid bilayer membranes. The transmembrane domains are essentially signal sequences that are found in the interior of the protein (that is, not at the N-terminus). They are not removed after translation. A single polypeptide can have several transmembrane domains passing back and forth through a membrane. During translation, the transmembrane domains are threaded through the ER membrane. The protein is then transported in vesicles to the Golgi apparatus and plasma membrane in the same manner as a secreted protein (see Figure 6). [For a protein in the plasma membrane, does the portion of the protein in the ER lumen end up facing the cytoplasm or the cellular exterior?19]
Additional functions of the rough ER include the initial post-translational modification of proteins. Although glycosylation (the addition of saccharides to proteins) is usually associated with the Golgi apparatus, some glycosylation occurs in the lumen of the ER. Disulfide bond formation also occurs in the ER lumen.
Two last notes about protein traffic throughout the cell: First, the default target for proteins that go through the secretory path is the plasma membrane. Targeting signals are needed if a protein going through that path needs to end up elsewhere (e.g., the Golgi, the ER, the lysosome). Second, proteins that are made in the cytoplasm but need to be sent to an organelle that is not part of the secretory path (e.g., the nucleus, mitochondria, or peroxisomes) require sequences called localization signals. The Table below summarizes protein traffic.
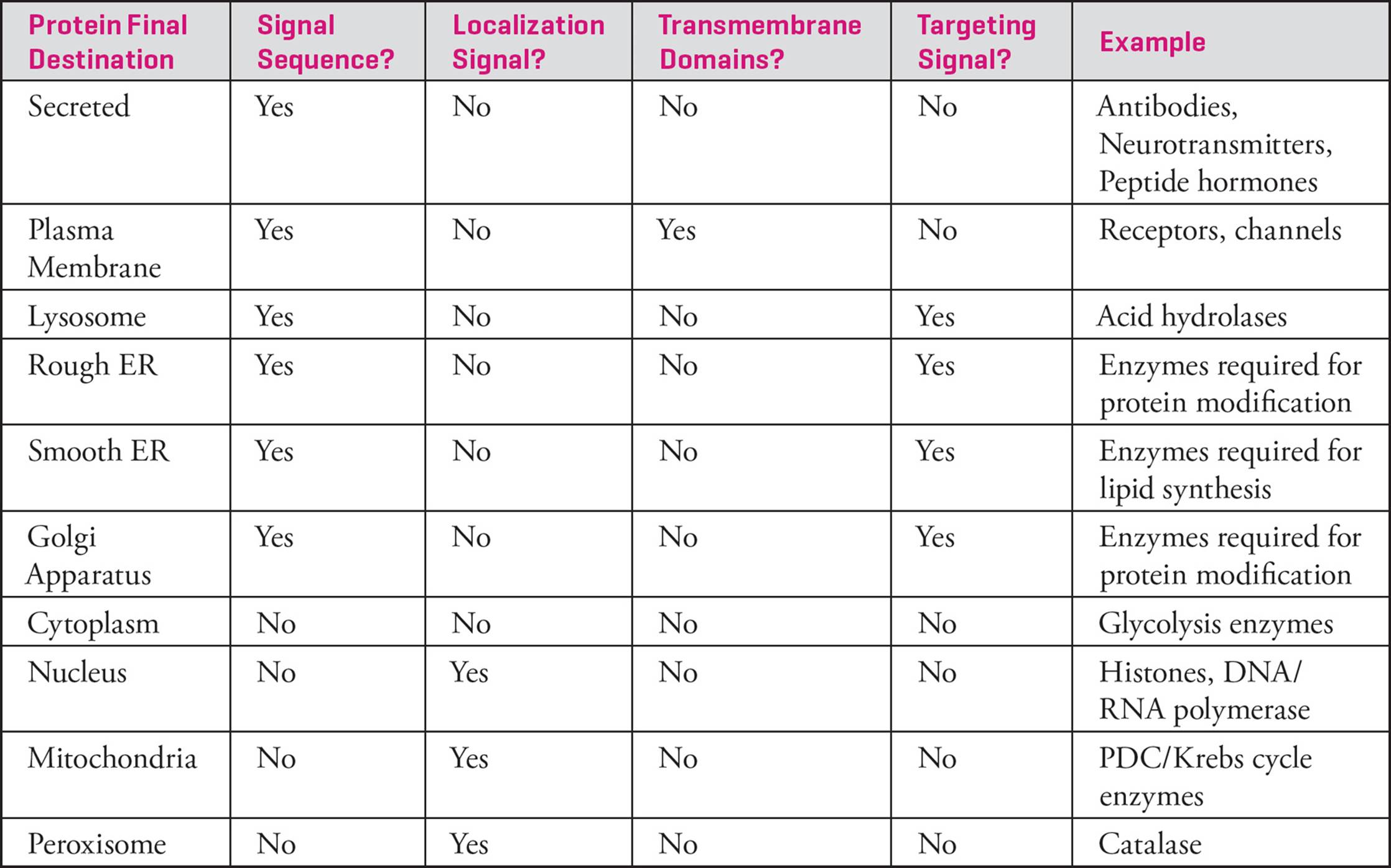
Table 2 Summary of Cellular Protein Traffic
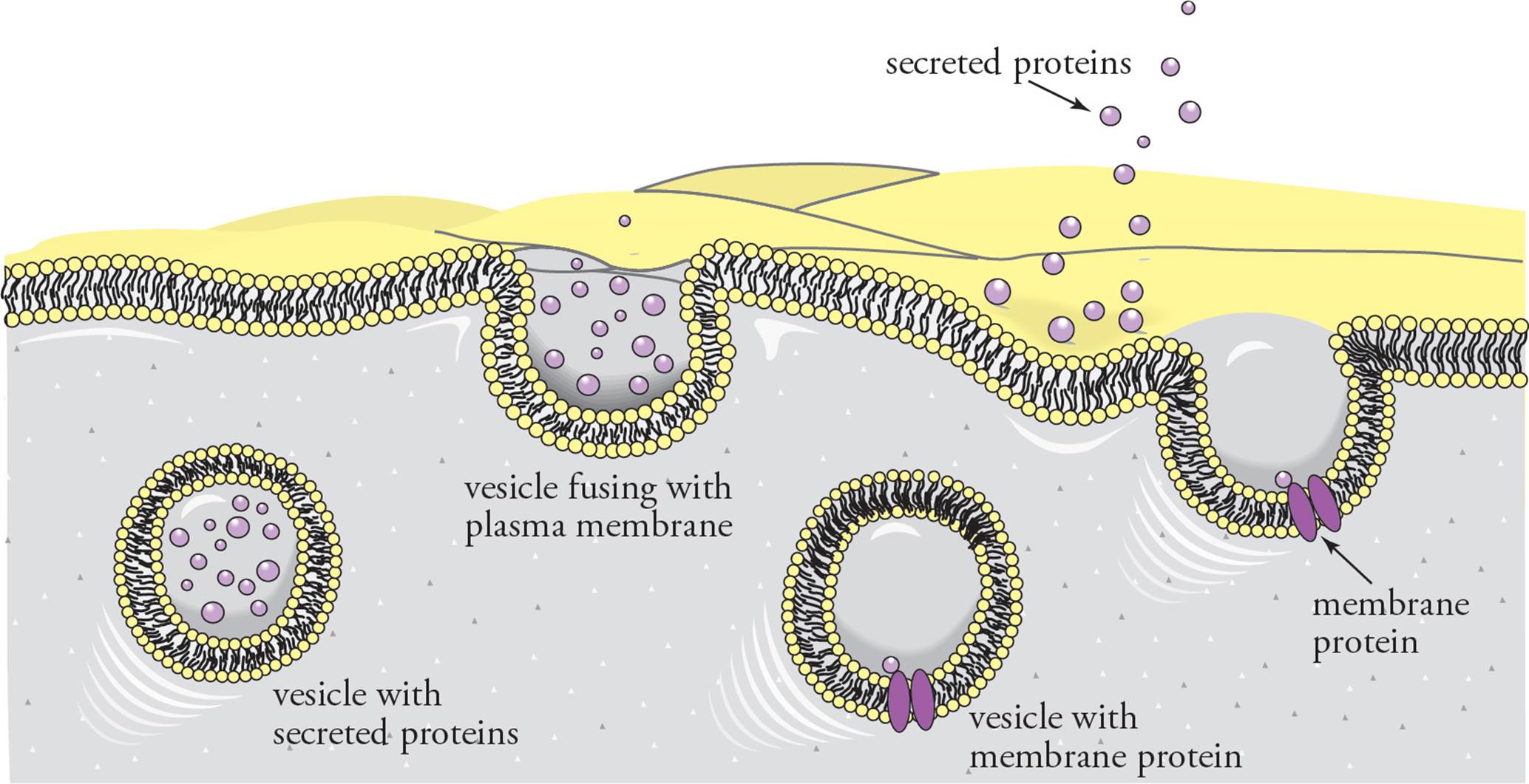
Figure 6 The Secretory Pathway—Secreted Proteins and Integral Membrane Proteins
• Disulfide bridges are found in extracellular proteins because the cytoplasm is a reducing environment that changes cystine to two cysteines. Given this fact, does it make sense that disulfide bridges are formed in the ER lumen?20
• Can mRNA coding for a protein destined to be embedded in the plasma membrane associate with rough ER prior to the initiation of translation?21
The Golgi Apparatus
The Golgi apparatus is a group of membranous sacs stacked together like collapsed basketballs (see Figure 7). It has the following functions: 1) Modification of proteins made in the RER; especially important is the modification of oligosaccharide chains. 2) Sorting and sending proteins to their correct destinations. 3) The Golgi also synthesizes certain macromolecules, such as polysaccharides to be secreted.
The vesicle traffic to and from the Golgi apparatus is mostly unidirectional; the membrane-bound or secreted proteins which are to be sorted and modified enter at one defined region and exit at another. (Traffic is said to be mostlyunidirectional because on occasion, proteins that are supposed to reside in the ER accidentally escape, and must be returned to the ER from the Golgi. This is called “retrograde traffic.”) Each region of the Golgi has different enzymes and a different microscopic appearance. The portion of the Golgi nearest the rough ER is called the cis stack, and the part farthest from the rough ER is the trans stack. The medial stack is in the middle.22 Vesicles from the ER fuse with the cis stack. The proteins in these vesicles are then modified and transferred to the medial stack, where they are further modified before passing to the trans stack. Proteins leave the Golgi at the trans face in transport vesicles. [If vesicle fusion with the cis Golgi was inhibited, could plasma membrane proteins still reach the cell surface?23] The route taken by a protein is determined by signals within the protein that determine which vesicle a protein is sorted into in the trans Golgi.
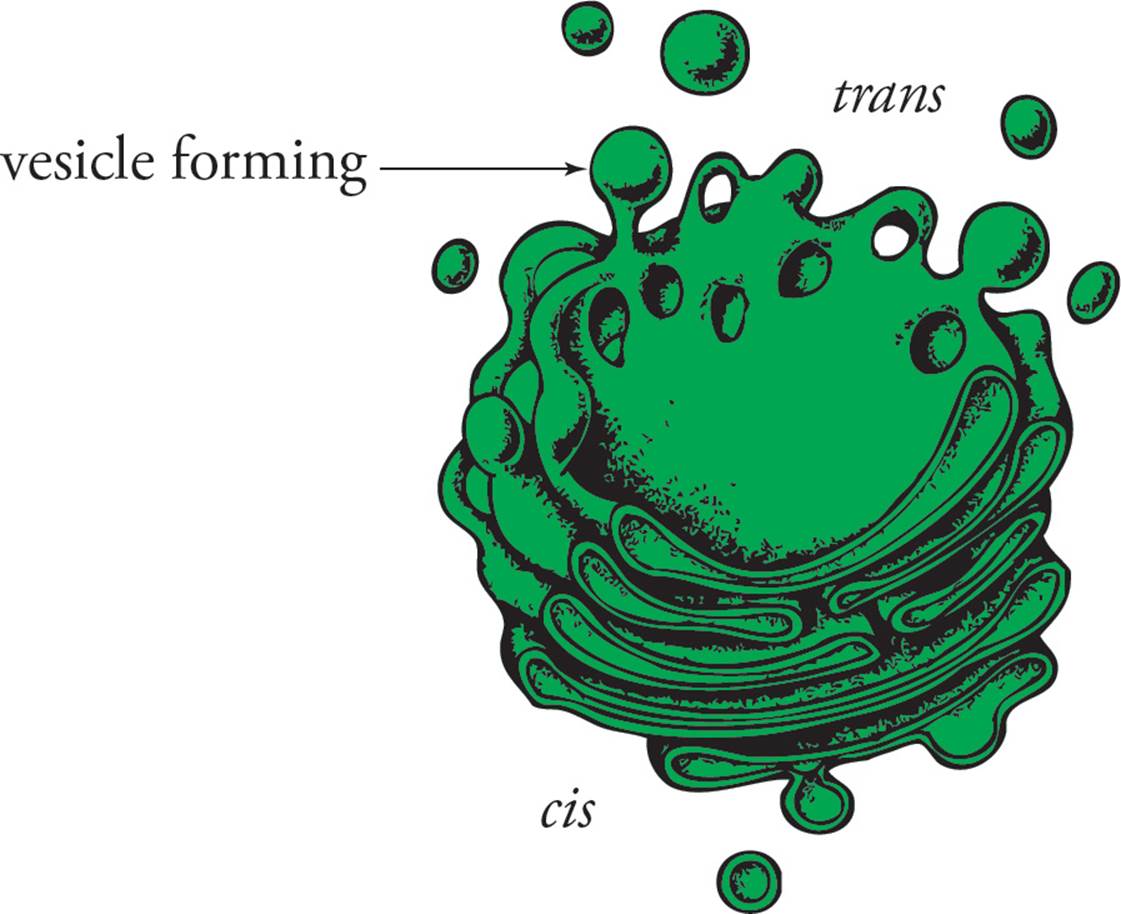
Figure 7 The Golgi Apparatus
When a vesicle moves from the trans Golgi toward the cell surface, it fuses with the cell membrane. As a result, the contents of the vesicle are released into the extracellular environment in a process termed exocytosis. Alternatively, if the vesicle contains proteins anchored to its membrane, these proteins will remain attached to the cell as cell-surface proteins. Some proteins are sent in vesicles from the Golgi immediately to the cell surface, in the constitutive secretory pathway. Constitutive connotes continuous or unregulated. In contrast, specialized secretory cells (such as pancreatic cells, B-cells of the immune system, etc.) store secretory proteins in secretory vesicles and release them only at certain times, usually in response to a change in (or signal from) the extracellular environment. This is a regulated secretory pathway.
Lysosomes
Lyse means cut. The lysosome is a membrane-bound organelle that is responsible for the degradation of biological macromolecules by hydrolysis. Lysosome proteins are made in the RER, modified in the Golgi, and released in their final form from the trans face of the Golgi. Organelles such as mitochondria that have been damaged or are no longer functional may be degraded in lysosomes in a process termed autophagy (self-eating). Lysosomes also degrade large particulate matter engulfed by the cell by phagocytosis (cell eating). For example, macrophages of the immune system engulf bacteria and viruses. The particle or microorganism ends up in a phagocytic vesicle, which will fuse with a lysosome. Finally, crinophagy refers to lysosomal digestion of unneeded (excess) secretory products. After hydrolysis, the lysosome will release molecular building blocks into the cytoplasm for reuse.
The enzymes responsible for degradation in lysosomes are called acid hydrolases. This name reflects the fact that these enzymes only hydrolyze substrates when they are in an acidic environment. This is a safety mechanism. The pH of the lysosome is around 5, so the acid hydrolases are active. But the pH of the cytoplasm is 7.4. If a lysosome ruptures, its enzymes will not damage the cell because the acidic fluid will be diluted, and the acid hydrolases will be inactivated. However, if many lysosomes rupture at once, the cell may be destroyed.
Peroxisomes
Peroxisomes are small organelles that perform a variety of metabolic tasks. The peroxisome contains enzymes that produce hydrogen peroxide (H2O2) as a by-product. They are essential for lipid breakdown in many cell types. In the liver they assist in detoxification of drugs and chemicals. H2O2 is a dangerous chemical, but peroxisomes contain an enzyme called catalase which converts it to H2O + O2. Separating these activities into the peroxisomes protects the rest of the cell from damage by peroxides or oxygen radicals.
7.3 THE PLASMA MEMBRANE
The evolution of life most likely began with a separation of “inside” from “outside.” Once this had occurred, processes in the cell could increase their orderliness despite the entropic chaos of the surroundings. An alternate hypothesis is that life began with self-replicating RNA floating free in the ocean. As it grew more complex, this early genome would require protection. In any case, the separation of the cytoplasm from the extracellular environment was a major milestone in evolution. Bacteria, plants, and fungi accomplish this by forming a cell membrane and a cell wall (made of peptidoglycan, cellulose, and chitin, respectively). Eukaryotic animal cells have no cell wall and thus rely on the cell membrane as the only boundary between inside and outside. And they must devise another means of structural support: just as chordates have a bony endoskeleton instead of the primitive exoskeleton arthropods have, animal cells rely on an internal cytoskeleton instead of an external cell wall. Further problems arise in multicellular eukaryotes. Not only must each cell maintain its structural integrity, but it must also interact with its neighbors in an organized fashion. In the following discussion, we will study how each of these goals is accomplished.
Membrane Structure
All of the membranes of the cell are composed of lipid bilayer membranes. The three most common lipids in eukaryotic membranes are phospholipids, glycolipids, and cholesterol, of which phospholipids are the most abundant. An example of a phospholipid is phosphatidyl choline (see Figure 8) with two long hydrophobic fatty acids esterified to glycerol, along with a charged phosphoryl choline group. Thus, phospholipids have portions that are distinctly hydrophilic and hydrophobic. Glycolipids, with fatty acids groups and carbohydrate side chains, also have hydrophilic and hydrophobic regions. When fatty acids or phospholipids are mixed with water, they spontaneously arrange themselves with the hydrophobic tails facing the interior to avoid contact with water and the hydrophilic regions facing outward toward water (see Figure 9). Fatty acids form small micelles, but, due to steric hindrance, phospholipids arrange themselves spontaneously into lipid bilayer membranes. Since the lipid bilayer is the lowest energy state for these molecules, the bilayer membrane can reseal and repair itself if a small portion of membrane is removed. [Does the formation of a lipid bilayer when phospholipids are mixed with water have a positive or a negative ∆G (change in free energy)?24]
The interior of the lipid bilayer membrane is very hydrophobic, with water largely excluded. Hydrophilic molecules such as ions, carbohydrates, and amino acids are not soluble in this environment, making the membrane a barrier to the passage of these molecules. Nonpolar molecules such as CO2, O2, and steroid hormones can cross the membrane easily. Water can also pass through the membrane but does so through specialized protein channels.
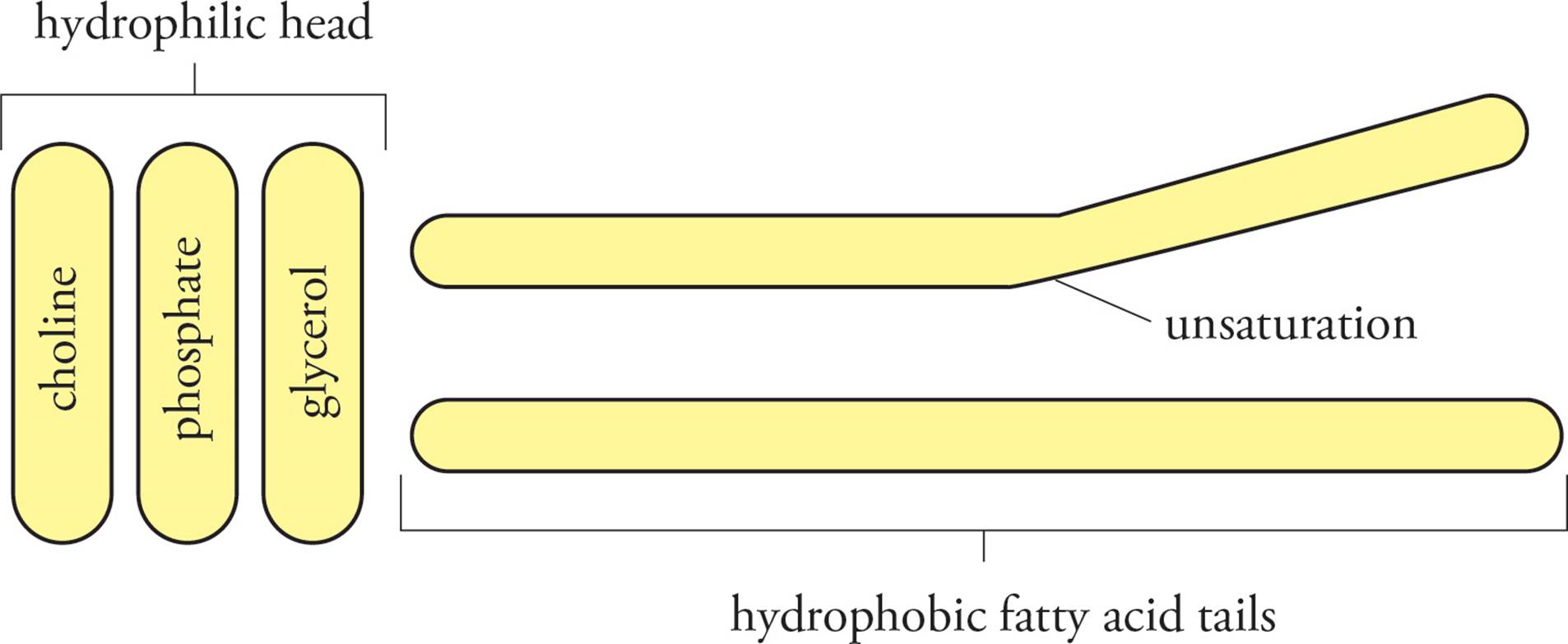
Figure 8 Phosphatidyl Choline, a Phospholipid
• Which one of the following statements best describes the physical characteristics of phospholipids?25
A) Negatively charged at pH 7 and therefore entirely hydrophilic
B) Hydrophobic
C) Partially hydrophilic and partially hydrophobic
D) Positively charged at pH 7 and therefore entirely hydrophilic
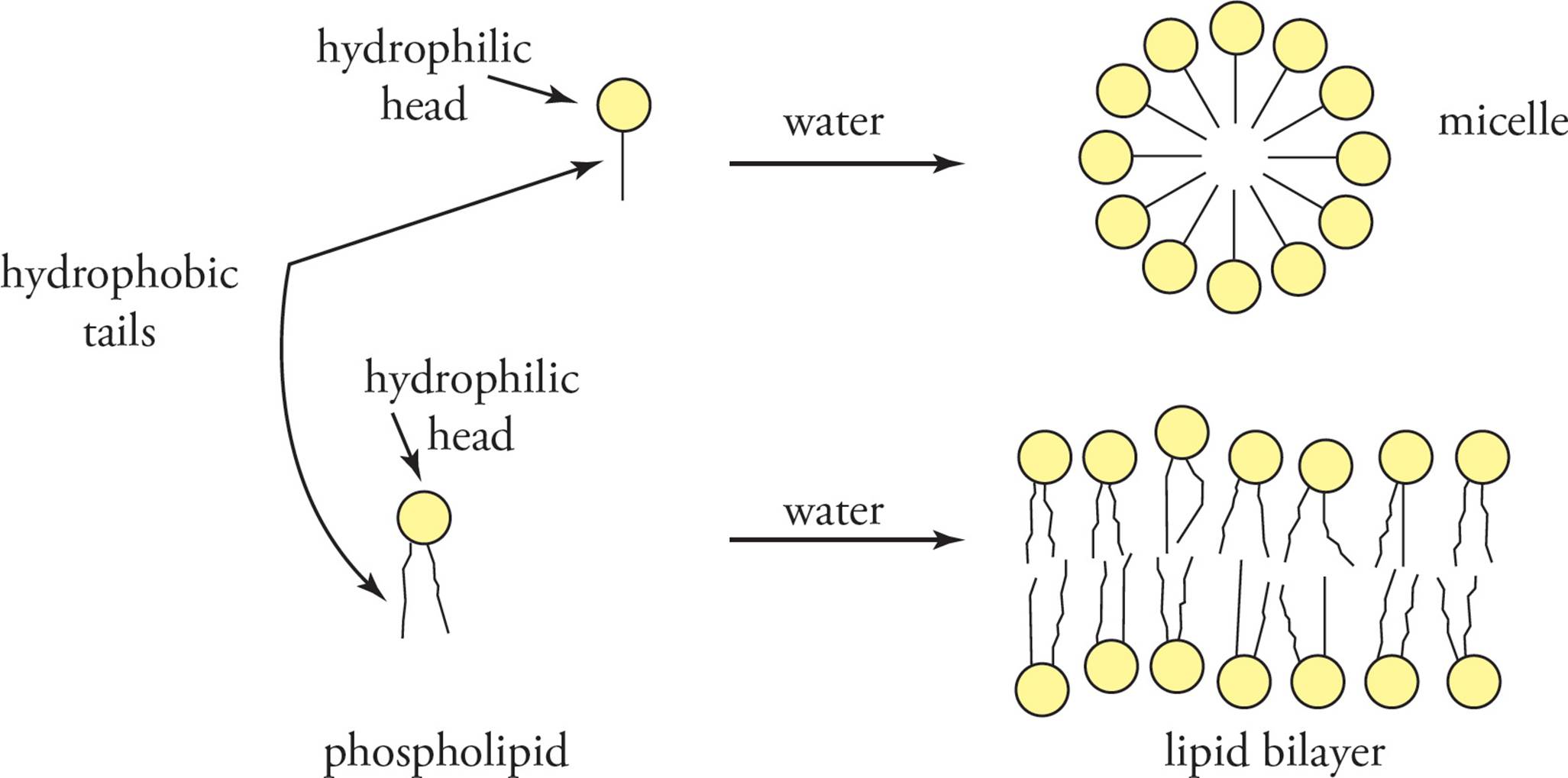
Figure 9 Lipid Behavior in an Aqueous Solvent
In addition to lipids, proteins are a major component of membranes. In some cases, such as the mitochondrial inner membrane, there is a higher protein than lipid concentration. Some proteins act to mediate interactions of the cell with other cells. Other proteins called cell-surface receptors bind extracellular signaling molecules such as hormones and relay these signals into the cell so that it can respond accordingly. Channel proteins selectively allow ions or molecules to cross the membrane. Each of these types of membrane protein is discussed below.
In general, membrane proteins are classified as peripheral or integral (see Figure 10). Integral membrane proteins are actually embedded in the membrane, held there by hydrophobic interactions. Membrane-crossing regions are called transmembrane domains (see Figure 11). Integral membrane proteins may have a complex pattern of transmembrane domains and portions not within the membrane. [At which point in the secretory pathway would the insertion of transmembrane domains into the membrane occur?26] Peripheral membrane proteins are not embedded in the membrane at all, but rather are stuck to integral membrane proteins, held there by hydrogen bonding and electrostatic interactions.
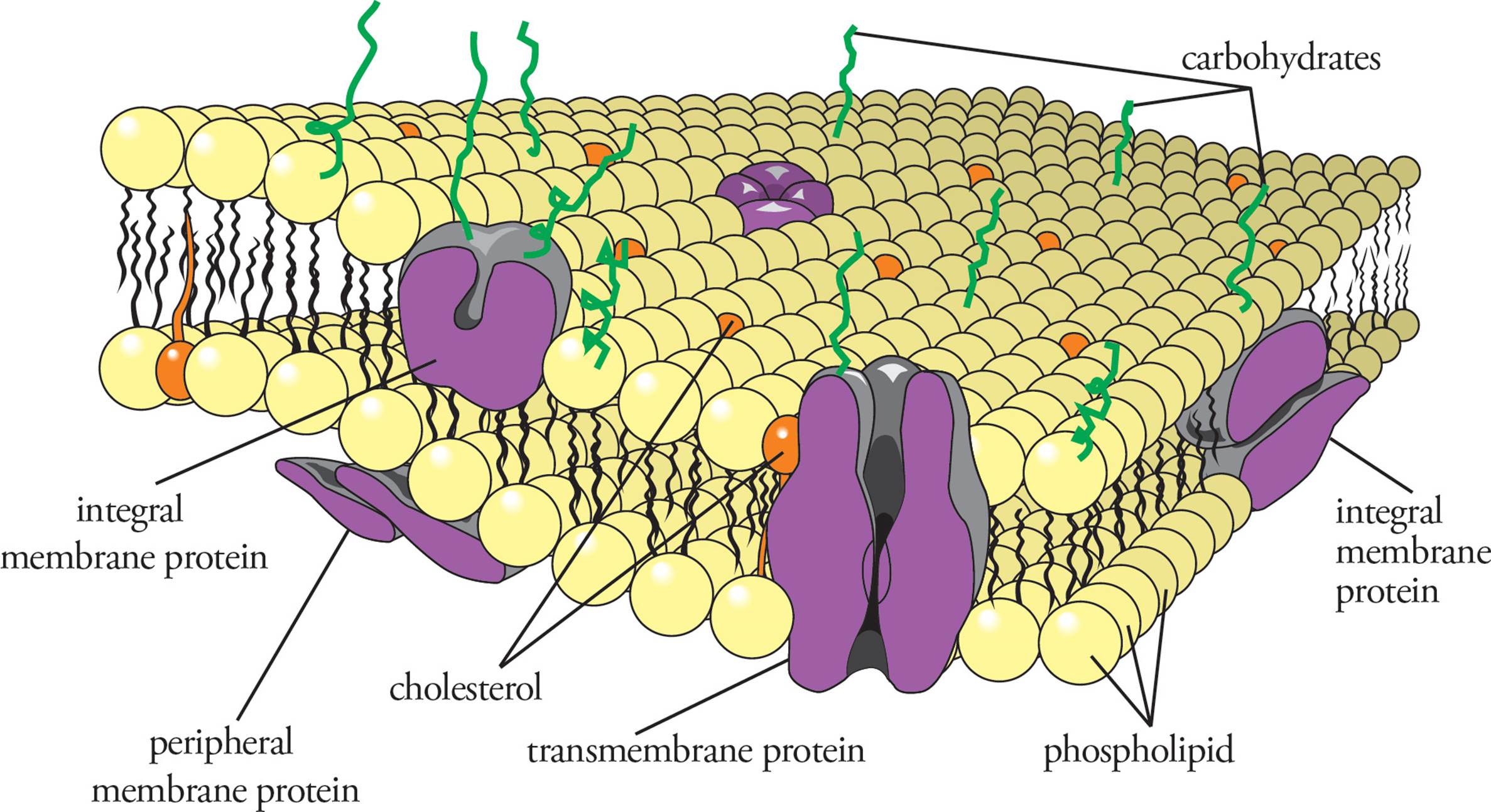
Figure 10 Membrane Proteins
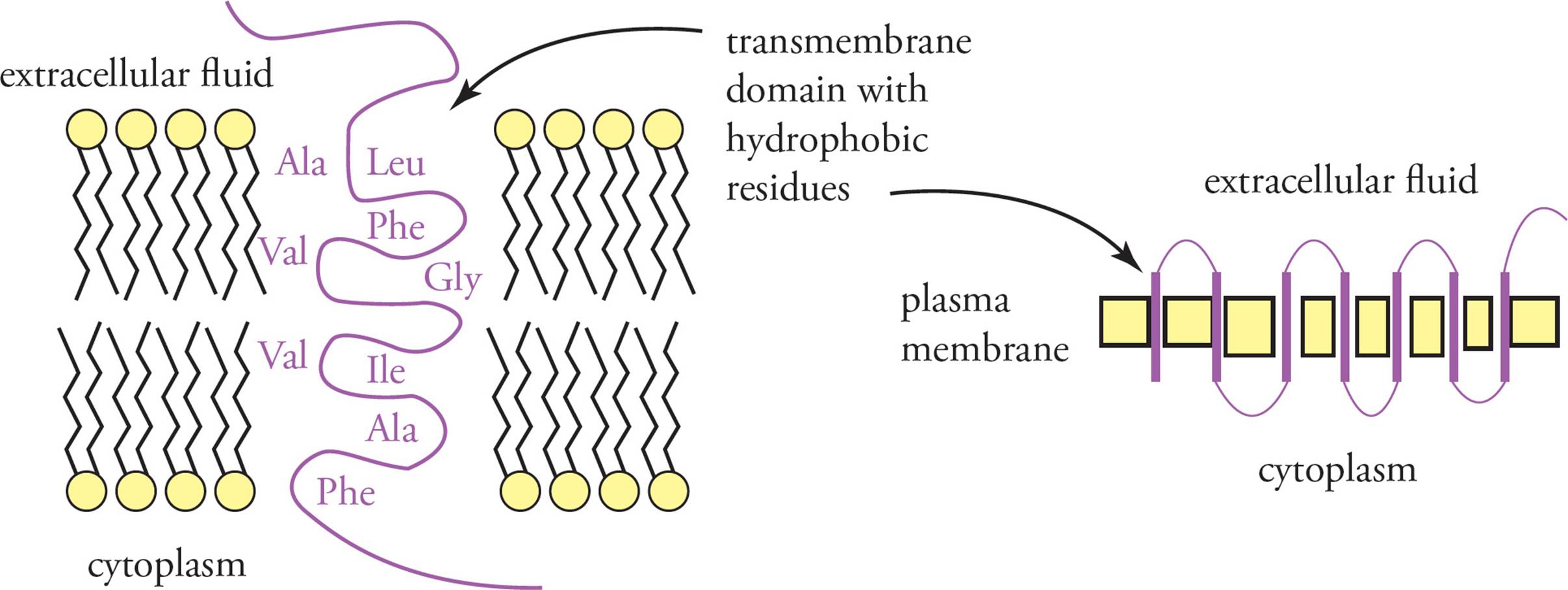
Figure 11 Transmembrane Domains
The current understanding of membrane dynamics is termed the fluid mosaic model, because the membrane is seen as a mosaic of lipids and proteins which are free to move back and forth fluidly. According to this model, lipids and proteins are free to diffuse laterally, in two-dimensions, but are not free to flip-flop. Phospholipid head groups and hydrophilic protein domains are restricted from entering the hydrophobic membrane interior just as hydrophilic molecules in the extracellular space are. Hence the membrane is said to have polarity. This just means that the inside face and the outside face remain different. We have already discussed one such difference: all glycosylations are found on the extracellular face. So the “fluid” in “fluid mosaic” means that things are free to move back and forth, but in two dimensions only. One exception is that some proteins are anchored to the cytoskeleton and thus cannot move in any direction.
• Phospholipids can be covalently attached to a fluorescent tag and then integrated into a lipid bilayer. If one cell has a red fluorescent tagged lipid in its plasma membrane and another cell has a green fluorescent tagged lipid in its membrane, what will happen if the two cells are fused together?27
The fluidity of a membrane is affected by the composition of lipids in the membrane (see Figure 12). The hydrophobic van der Waals interactions between the fatty acid side chains are a major determinant of membrane fluidity. Saturated fatty acids, lacking any double bonds, have a very straight structure and pack tightly in the membrane, with strong van der Waals forces between side chains. Unsaturated fatty acids, with one or more double bonds, have a kinked structure and pack in the membrane interior more loosely. Cholesterol also plays a key role in maintaining optimal membrane fluidity by fitting into the membrane interior. [If the percentage of unsaturated fatty acids in a membrane is increased, will membrane fluidity increase or decrease at body temperature?28]
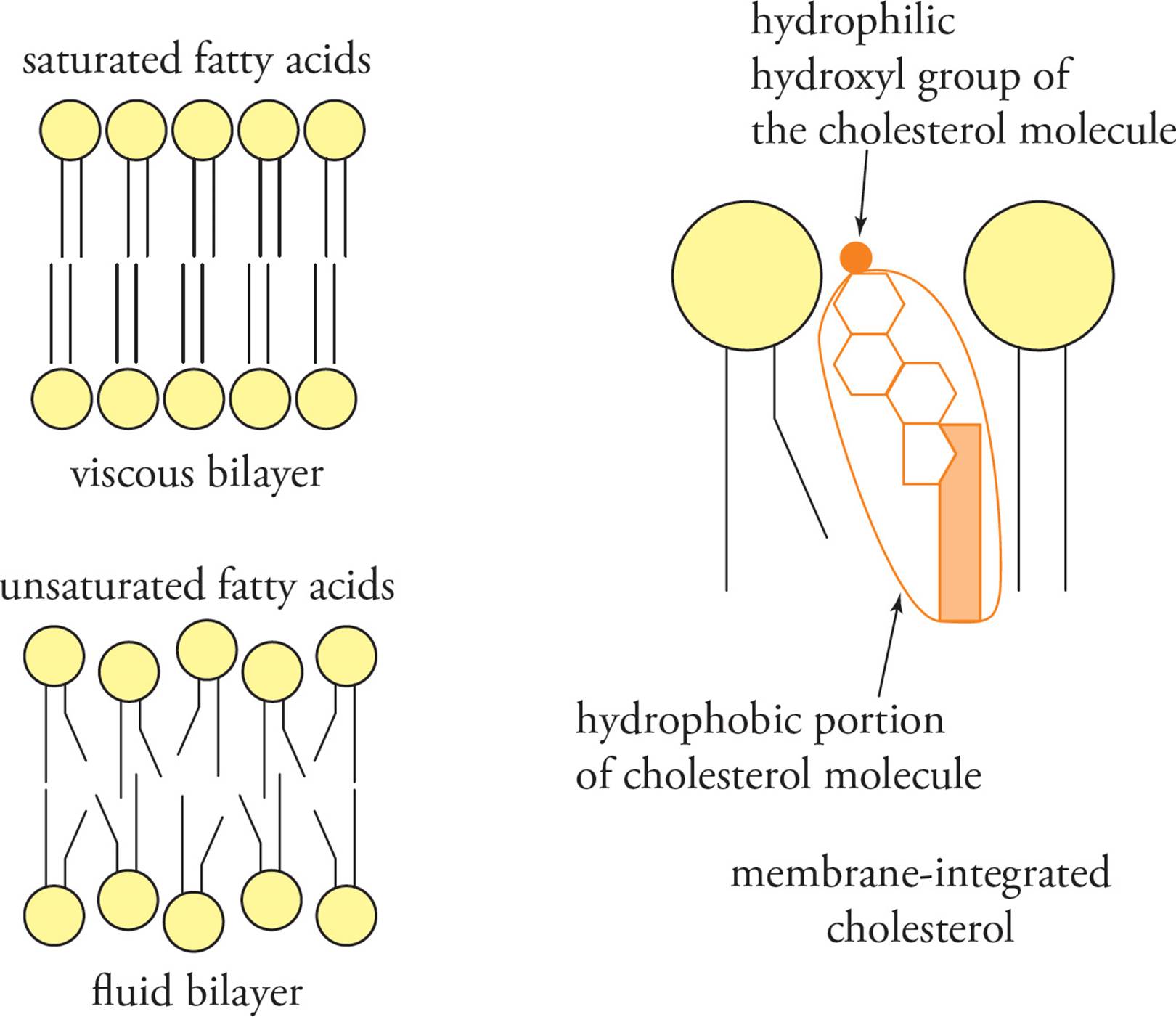
Figure 12 Factors Affecting Membrane Fluidity
7.4 TRANSMEMBRANE TRANSPORT
The cell requires membranes to act as barriers to diffusion but also requires the transport of many different substances across membranes. Integral membrane proteins transport material through membranes that cannot diffuse on their own across membranes. Transport across a membrane can be either passive (does not require cellular energy) or active (requires cellular energy). Before we discuss movements across membranes, let’s review basic rules about concentration, ionizability, colligative properties, and diffusion and osmosis.
Concentration Measurements
Molarity (M) expresses the concentration of a solution in terms of moles of solute per volume (in liters) of solution:
Molarity (M) = 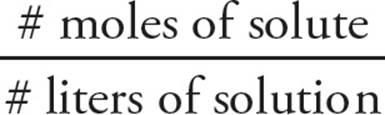
Concentration is denoted by enclosing the solute in brackets. For instance, “[Na+] = 1.0M” indicates a solution whose concentration is equivalent to 1 mole of sodium ions per liter of solution.
Molality (m) expresses concentration in terms of moles of solute per mass (in kilograms) of solvent:
Molarity (m) = 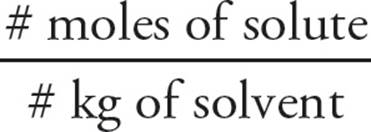
Molality is particularly useful when measuring properties that involve temperature because, unlike molarity, molality does not change with temperature. And, since a liter of water has a mass of one kilogram, the molar and molal concentrations of dilute aqueous solutions are nearly the same. This is particularly true in biological systems, where the volume (essentially a cell) is very small and the solvent is always water.
Mole fraction simply expresses the fraction of moles of a given substance (which we’ll denote here by S) relative to the total moles in a solution:
mole fraction of S = Xs = 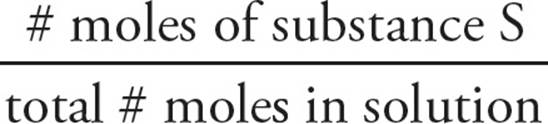
Mole fraction is a useful way to express concentration when more than one solute is present.
Electrolytes
When ionic substances dissolve, they dissociate into ions. Free ions in a solution are called electrolytes because the solution can conduct electricity. Some salts dissociate completely into individual ions, while others only partially dissociate (that is, a certain percentage of the ions will remain paired, sticking close to each other rather than being independent and fully surrounded by solvent). Solutes that dissociate completely (like ionic substances) are called strong electrolytes, and those that remain ion-paired to some extent are called weak electrolytes. (Covalent compounds that don’t dissociate into ions are nonelectrolytes.) Solutions of strong electrolytes are better conductors of electricity than those of weak electrolytes.
Different ionic compounds will dissociate into different numbers of particles. Some won’t dissociate at all, and others will break up into several ions. The van’t Hoff (or ionizability) factor (i) tells us how many ions one unit of a substance will produce in a solution. For example,
• C6H12O6 is non-ionic, so it does not dissociate. Therefore, i = 1. (Note: The van’t Hoff factor for almost all biomolecules—hormones, proteins, steroids, etc.—is 1.)
• NaCl dissociates into Na+ and Cl–. Therefore, i = 2.
• HNO3 dissociates into H+ and NO3–. Therefore, i = 2.
• CaCl2 dissociates into Ca2+ and 2 Cl–. Therefore, i = 3.
Colligative Properties
Colligative properties depend on the number of solute particles in the solution rather than the type of particle. For example, when any solute is dissolved into a solvent, the boiling point, freezing point, and vapor pressure of the solution will be different from those of the pure solvent. For colligative properties, the identity of the particle is not important. That is, for a 1 M solution of any solute, the change in a colligative property will be the same no matter what the size, type, or charge of the solute particles. Remember to consider the van’t Hoff factor when accounting for particles: One mole of sucrose (i = 1) will have the same number of particles in solution as 0.5 mol of NaCl (i = 2), and therefore will have the same effect on a colligative property. Thus, we can consider the effective concentration to be the product iM (or im); this is the concentration of particles present.
The four colligative properties we’ll study for the MCAT are vapor-pressure depression, boiling-point elevation, freezing-point depression, and osmotic pressure.
Vapor-Pressure Depression
Think about being at the ocean or a lake in the summer. The air is always more humid (moist) than in the middle of a parking lot. Why? Because some of the water molecules gain enough energy to get into the gas phase, so we see a dynamic equilibrium setup between the molecules in the liquid phase and the molecules in the gas (vapor) phase.
Vapor pressure is the pressure exerted by the gaseous phase of a liquid that evaporated from the exposed surface of the liquid. The weaker a substance’s intermolecular forces, the higher its vapor pressure and the more easily it evaporates. For example, if we compare diethyl ether, H5C2OC2H5, and water, we notice that while water undergoes hydrogen bonding, diethyl ether does not, so despite its greater molecular mass, diethyl ether will vaporize more easily and have a higher vapor pressure than water. Easily vaporized liquids—liquids with high vapor pressure—like diethyl ether are said to be volatile.
Now let’s think about what happens to vapor pressure when the liquid contains a dissolved solute. The solute molecules are attached to solvent molecules and act as “anchors.” As a result, more energy is required to enter the gas phase since the solvent molecules need to break away from their interactions with the solute before they can enter the gas phase. In fact, the boiling point of a liquid is defined as the temperature at which the vapor pressure of the solution is equal to the atmospheric pressure over the solution. Thus, at sea level, where the atmospheric pressure is 760 torr, the solution must have a vapor pressure of 760 torr in order to boil. Adding more solute to the same solution will decrease its vapor pressure. Boiling will still take place when vapor pressure is 760 torr, but more heat will have to be supplied to reach this vapor pressure, and thus the solution will boil at a higher temperature. For example, salted water (say, for cooking spaghetti) boils at a higher temperature than unsalted water.
Boiling-Point Elevation
When a liquid boils, the molecules in the liquid acquire enough energy to overcome the intermolecular forces and break free into the gas phase. The liquid molecules escape as a vapor at the surface between the liquid and air. But what happens when a non-volatile solute is added to the liquid? As described before, the solute particles are attached to solvent molecules and act as “anchors.” As a result, more energy is required since you not only have to convert the solvent into the gas phase, but you first have to break the interaction with the solute. What happens to the boiling point? In order for the molecules to escape, they need more energy than they did without the solute. This translates into an elevation of the boiling point. The increase in boiling point is directly related to the number of particles in solution and the type of solvent. For a given solvent (again, in biological systems this is always water), the more solute particles, the greater the boiling-point elevation. Also, you have to consider that some compounds dissociate when they dissolve, so the equation for boiling-point elevation includes the van’t Hoff factor, i:
Boiling-Point Elevation
∆Tb = kbim
In this equation, kb is the solvent’s boiling-point elevation constant, i is the solute’s van’t Hoff factor, and m is the molal concentration of the solution. For water, kb ≈ 0.5°C / m.
Freezing-Point Depression
What happens when we add a solute to a liquid, then try to freeze the solution? Solids are held together by attractive intermolecular forces. During freezing, the molecules in a liquid will assemble into an orderly, tightly packed array. However, the presence of solute particles will interfere with efficient arrangement of the solvent molecules into a solid lattice. As a result, a liquid will be less able to achieve a solid state when a solute is present, and the freezing point of the solution will decrease. (Or, equivalently, the melting point of a solid containing a solute is decreased.) The good news is that the formula for freezing-point depression has exactly the same form as the formula for boiling-point elevation, except that the temperature is going down instead of up (that is, the equation for freezing-point depression has a minus sign whereas the equation for boiling-point elevation has a plus sign).
Freezing-Point Depression
∆Tf = −kfim
In this equation, kf is the solvent’s freezing-point depression constant, i is the solute’s van’t Hoff factor, and m is the molal concentration of the solution. For water, kf ≈ 1.9°C/m.
• Addition of concentrated sulfuric acid to pure water will result in:29
A) vapor-pressure depression.
B) boiling-point elevation.
C) freezing-point depression.
D) all of the above.
Review of Diffusion and Osmosis
Diffusion is the tendency for liquids and gases to fully occupy the available volume (Figure 13). Particles in the liquid or gas phase are in constant motion, depending on temperature. If all particles are concentrated in one portion of a container, we have an orderly situation, which is unfavorable according to the second law of thermodynamics (law of entropy). The constant thermal motion of particles in the cell leads to their spreading out to occupy all available space, which maximizes entropy.30 A solute will always diffuse down its concentration gradient, which means from high to low concentration. Diffusion continues until the solute is evenly distributed throughout the available volume. At this point, movement of solute back and forth continues, but no net movement occurs.
Osmosis is a special type of diffusion in which solvent diffuses rather than solute (Figure 13). For example, if a chamber containing water and a chamber containing a solution of sucrose are connected directly, sucrose will diffuse throughout the entire volume until a uniform concentration is reached. However, if the two chambers are separated by a semipermeable membrane that allows water but not sucrose to cross, then diffusion of sucrose between the chambers cannot occur. In this case, osmosis draws water into the sucrose chamber to reduce the sucrose concentration as well as the volume in the water chamber. Ignoring gravity, water will flow into the sucrose chamber until the concentration is the same across the membrane. The plasma membrane of the cell is a semipermeable membrane that allows water—but not most polar solutes—to cross by osmosis. [If a cell is placed in a hypotonic solution (solute concentration lower than in the cell), what will happen to the cell?31]
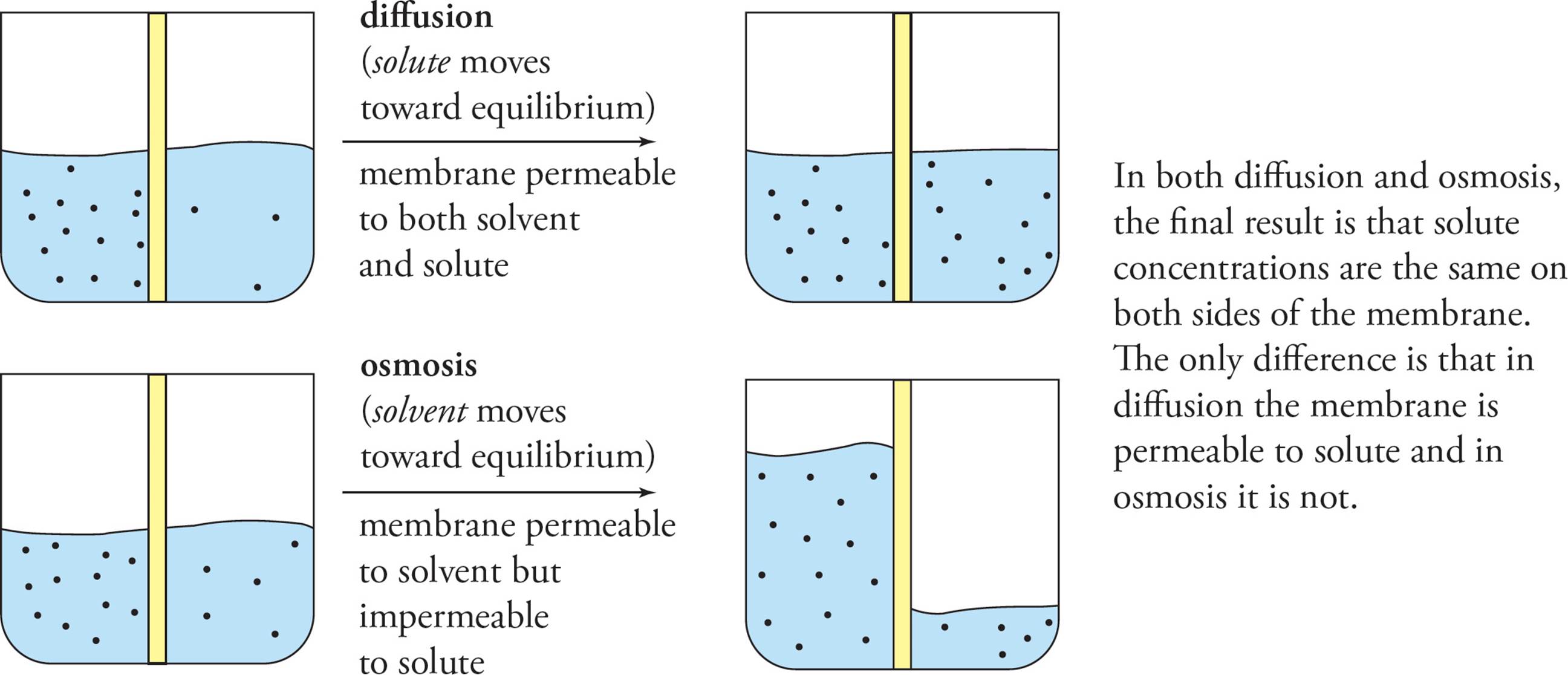
Figure 13 Diffusion and Osmosis
The term tonicity is used to describe osmotic gradients. If the environment is isotonic to the cell, the solute concentration is the same inside and outside. A hypertonic solution has more total dissolved solutes than the cell, a hypotonic solution has less. You may also hear the terms isoosmotic, hyperosmotic, and hypoosmotic. The tendency of water to move down its concentration gradient (into cells) can be a powerful force, able to cause cells to explode. This tendency (of water to move to where there are more particles) along with the inability of those particles to cross the membrane is what accounts for the difference in fluid levels in the beaker at the bottom right-hand corner of Figure 13. The large difference in fluid levels may be a rather extreme example, but it is conceptually accurate: Just as osmotic forces can cause a cell to rupture, they can overcome gravity, as shown.
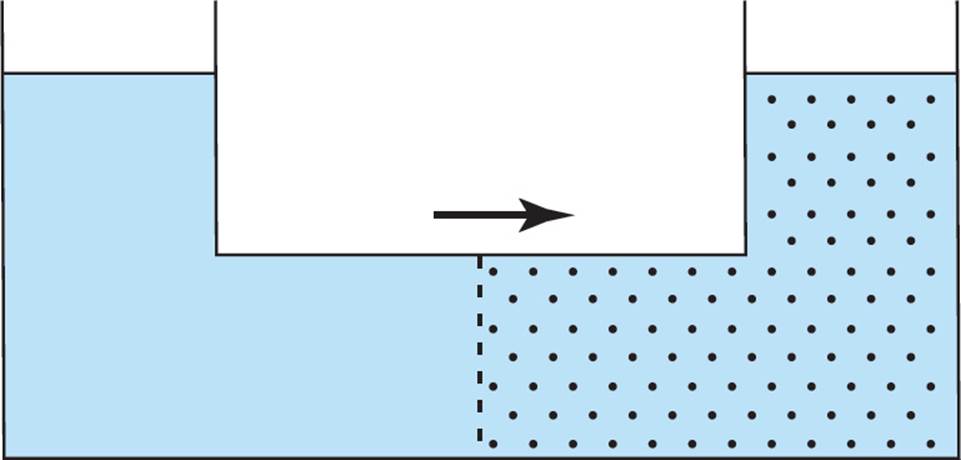
Osmotic Pressure
Osmosis describes the net movement of water across a semipermeable membrane from a region of low solute concentration to a region of higher solute concentration in an effort to dilute the higher concentration solution. The semipermeable membrane prohibits the transfer of solutes, but allows water to transverse through it. In the following figure, the net movement of water will be to the right:
Osmotic pressure (∏) can be defined as the pressure it would take to stop osmosis from occurring. If a pressure gauge were added to the same system, osmotic pressure could be measured.
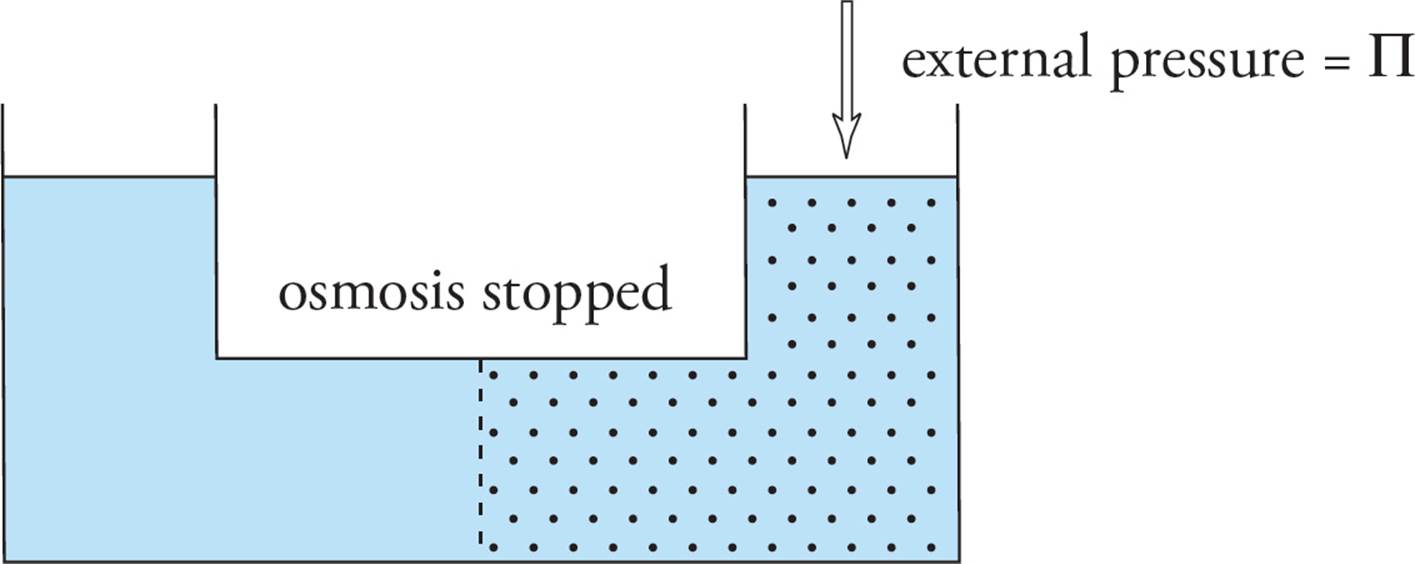
The osmotic pressure of a solution is given by the van’t Hoff equation:
∏ = MiRT
where ∏ is osmotic pressure in atm, M is the molarity of the solution, i is the van’t Hoff factor, R is the universal gas constant (0.0821 L-atm/K-mol), and T is the temperature in kelvins.
Again, changes in osmotic pressure are affected only by the number of particles in solution (taking into account the van’t Hoff factor), not by the identity of those particles.
Now let’s continue the discussion on movements across the cell membrane.
Passive Transport
Passive transport is a biochemical term that means diffusion. It refers to any thermodynamically favorable movement of solute across a membrane. Another way to phrase this is to say that passive transport is any movement of solute down a gradient. No energy is required since the concentration gradient drives movement of the solute. There are two types of passive transport: simple diffusion and facilitated diffusion.
Simple Diffusion
Simple diffusion is diffusion of a solute through a membrane without help from a protein. For example, steroid hormones are free to move back and forth across the membrane by simple diffusion as pushed by concentration gradients, thanks to their __.32
However, lipid bilayer membranes are impermeable to most solutes; that is one of the main functions of membranes. The plasma membrane is a barrier to the free movement of all large and/or hydrophilic solutes. Facilitated diffusion is the movement of a solute across a membrane, down a gradient, when the membrane itself (the pure lipid bilayer) is intrinsically impermeable to that solute. Specific integral membrane proteins allow material to cross the plasma membrane down a gradient in facilitated diffusion. For example, red blood cells require glucose, which they get from the bloodstream. However, glucose is a bulky hydrophilic molecule that cannot cross the RBC lipid bilayer. Instead, it must be shuttled across by a particular protein in the RBC plasma membrane. There are two well-characterized types of proteins which serve this sort of function: channel proteins and carrier proteins. Channels and carriers give the membrane its essential feature of selective permeability; permeability to some things despite impermeability to most things.
Facilitated Diffusion: Channels
Channel proteins in the plasma membrane allow material that cannot pass through the membrane by simple diffusion to flow through the plasma membrane down a concentration gradient. Channels do this by forming a narrow opening in the membrane surrounded by the protein. Channels are very selective in what passes through the opening in the membrane. There are many kinds of ion channels, each of which allows the passage of only one type of ion through the channel down a gradient (see Figure 14). All cells have potassium ion channels, for example, that allow only potassium (and not sodium) to flow through the plasma membrane down a gradient. Ion channels are said to be gated if the channel is open in response to specific environmental stimuli. A channel that opens in response to a change in the electrical potential across the membrane is called a voltage-gated ion channel. One that opens in response to binding of a specific molecule like a neurotransmitter is called a ligand-gated ion channel. The regulation of membrane potential by gated ion channels plays a key role in the nervous system. [Can ion channels move ions against an electrochemical gradient?33]
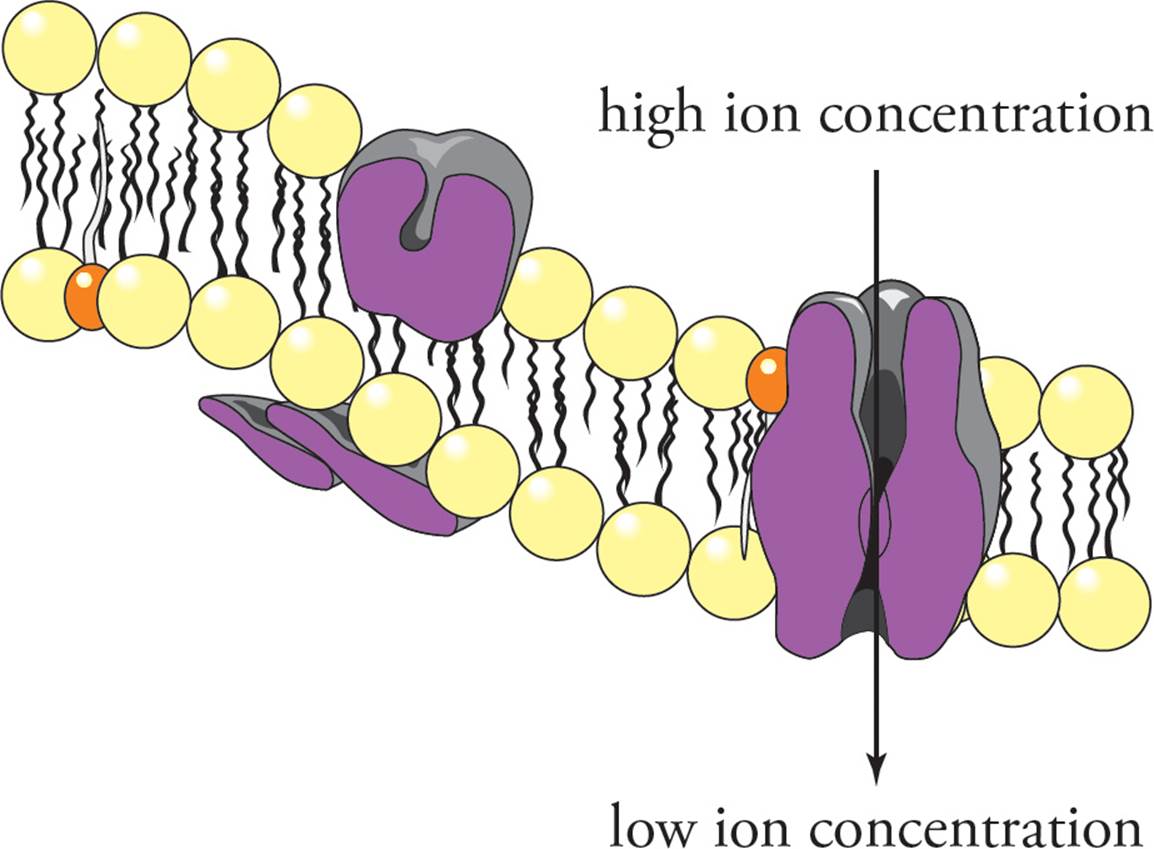
Figure 14 An Ion Channel
Facilitated Diffusion: Carriers
Carrier proteins also can transport molecules through membranes by facilitated diffusion, but they do so by a mechanism different from that of ion channels. Carrier proteins do not form a tunnel through membranes like ion channels do. Instead, carriers appear to bind the molecule to be transported at one side of the membrane and then undergo a conformational change to move the molecule to the other side of the membrane. Some carriers, called uniports, transport only one molecule across the membrane at a time (see Figure 15). Other carriers termed symports carry two substances across a membrane in the same direction. Antiports, on the other hand, carry two substances in opposite directions.
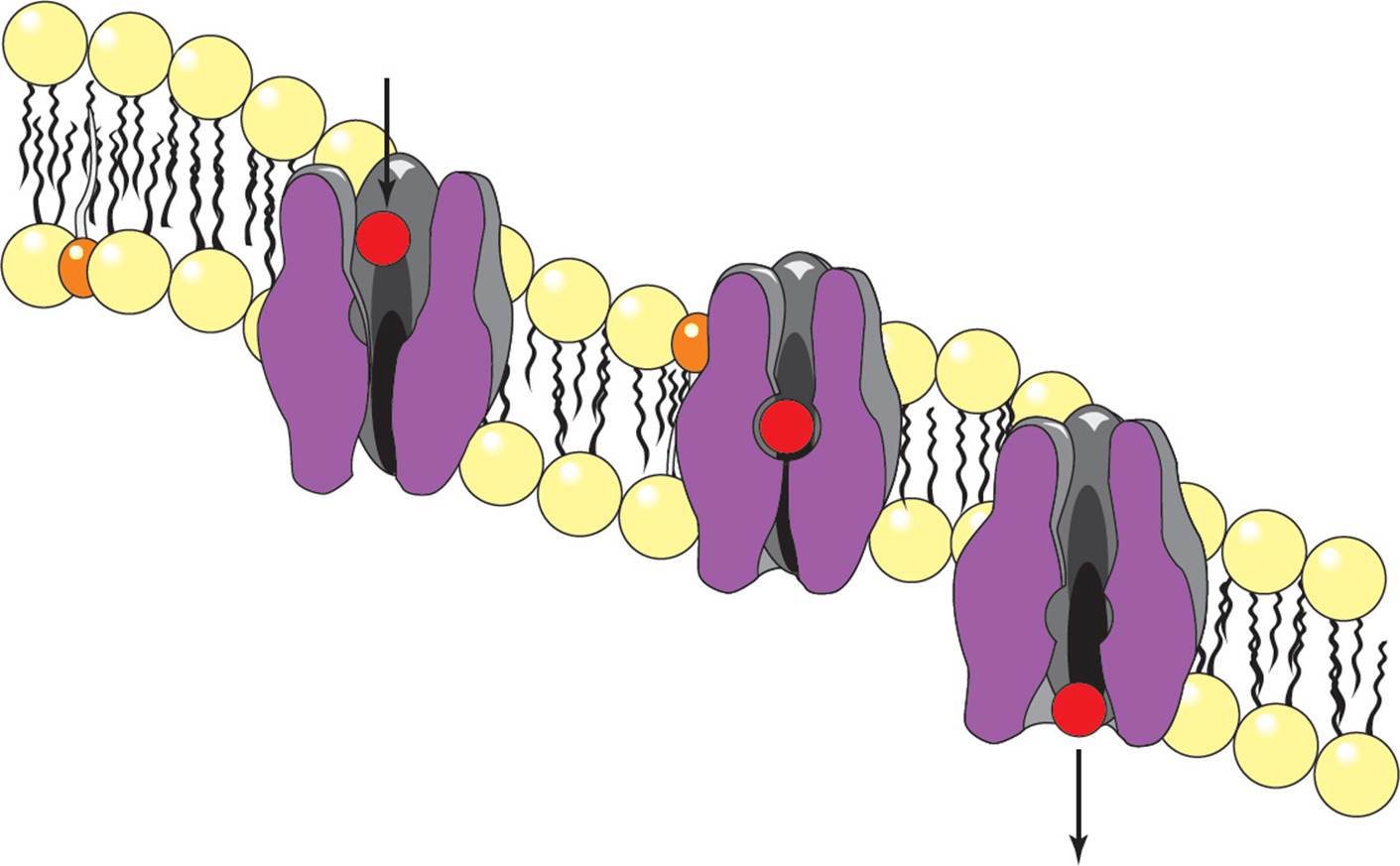
Figure 15 A Uniport
Pores and Porins
A pore is a tube through the membrane which is so large that it is not selective for any particular molecule. Rather, all molecules below a certain size may pass. (Also, a molecule which is just barely small enough to cross may not cross if it has the wrong charge on its surface.) Pores are formed by polypeptides known as porins. You are already familiar with several examples of pores. We have studied pores in the double nuclear membrane, the outer mitochondrial membrane, and the Gram-negative bacterial outer membrane. The eukaryotic plasma membrane does not have pores, because pores destroy the barrier function of the membrane, allowing solutes in the cytoplasm to freely diffuse out of the cell. [Are porins and ion channels found in the same membranes?34]
Kinetic Concerns
Simple diffusion can be distinguished from all forms of facilitated diffusion by the kinetics of the process. The rate of simple diffusion is limited only by the surface area of the membrane and the size of the driving force (gradient). Facilitated diffusion, however, depends on a finite number of integral membrane proteins. Hence, it exhibits saturation kinetics. Increasing the driving force for facilitated diffusion increases the rate of diffusion (the flux), but only to a point. Then all the transport proteins become saturated, and no further increase in flux is possible (Figure 16).
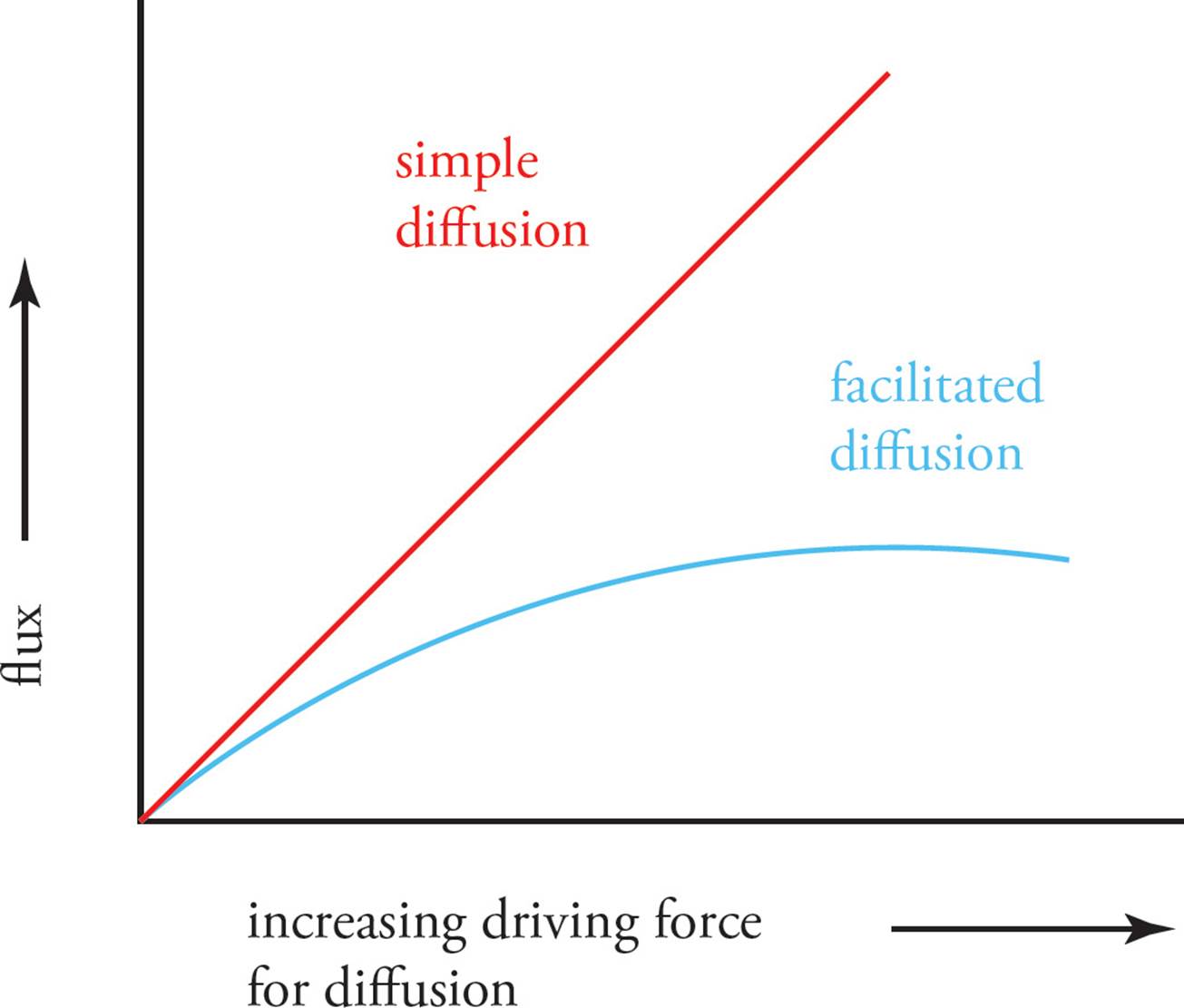
Figure 16 Saturation Kinetics of Facilitated Diffusion
Active Transport
Active transport is the movement of molecules through the plasma membrane against a gradient. Active transport requires energy input, since it is working against a gradient, and always involves a protein. Another way of saying that active transport requires energy input is to say that the transport process is coupled to a process which is thermodynamically favorable (∆G < 0). The gradient being pumped against is not necessarily just a concentration gradient, but for charged molecules, like ions, it can also involve electric potentials that form a combined electrochemical gradient that must be pumped against. The form of energy input used to drive movement of molecules against an electrochemical gradient varies. In primary active transport, the transport of a molecule is coupled to ATP hydrolysis. In secondary active transport, the transport process is not coupled directly to ATP hydrolysis. Instead, ATP is first used to create a gradient, then the potential energy in that gradient is used to drive the transport of some other molecule across the membrane. Since ATP is not used in the actual transport of the “other” molecule, the ATP use is described as indirect. For example, the transport of glucose into some cells is driven against the glucose concentration gradient by the cotransport of sodium ions down the sodium electrochemical gradient, previously established by an ATPase pump (see below). A common mechanism driving secondary active transport of many different molecules involves coupling transport to the flow of sodium ions down a gradient.
• If a protein moves sodium ions across the plasma membrane down an electrochemical gradient, what form of transport is this?35
A) Simple diffusion
B) Facilitated diffusion
C) Primary active transport
D) Secondary active transport
The Na+/K+ ATPase and the Resting Membrane Potential
The Na+/K+ ATPase is a transmembrane protein in the plasma membrane of all cells in the body. The activity provided by this protein is to pump 3 Na+ out of the cell, 2 K+ into the cell, and to hydrolyze one ATP to drive the pumping of these ions against their gradients (Figure 17). [The pumping of sodium and potassium by the Na+/K+ ATPase is an example of what form of transport?36] The sodium which is pumped out of the cell stays outside, since the plasma membrane is impermeable to sodium ions. Some of the potassium ions which are pumped into the cell are able to leak back out, however, through potassium leak channels. Potassium flows down its concentration gradient out of the cell through leak channels. The movement of ions out of the cell helps the cell to maintain osmotic balance with its surroundings. As potassium leaves the cell through the leak channels, the movement of positive charge out of the cell creates an electric potential across the plasma membrane with a net negative charge on the interior of the cell. This potential created by the Na+/K+ ATPase is known as the resting membrane potential. (The resting membrane potential will be examined again in Chapter 7 in relation to action potentials in neurons). The concentration gradient of high sodium outside of the cell established by the Na+/K+ ATPase is the driving force behind secondary active transport of many different molecules, including sugars and amino acids. To summarize, the activity of the Na+/K+ ATPase is important in three ways:
1) To maintain osmotic balance between the cellular interior and exterior.
2) To establish the resting membrane potential.
3) To provide the sodium concentration gradient used to drive secondary active transport.
• If an inhibitor of Na+/K+ ATPase is added to cells, which of the following may occur?37
A) The cell will shrink and lose water.
B) The interior of the cell will become less negatively charged.
C) Secondary active transport processes will compensate for the loss of primary active transport.
D) The cell will begin to proliferate.
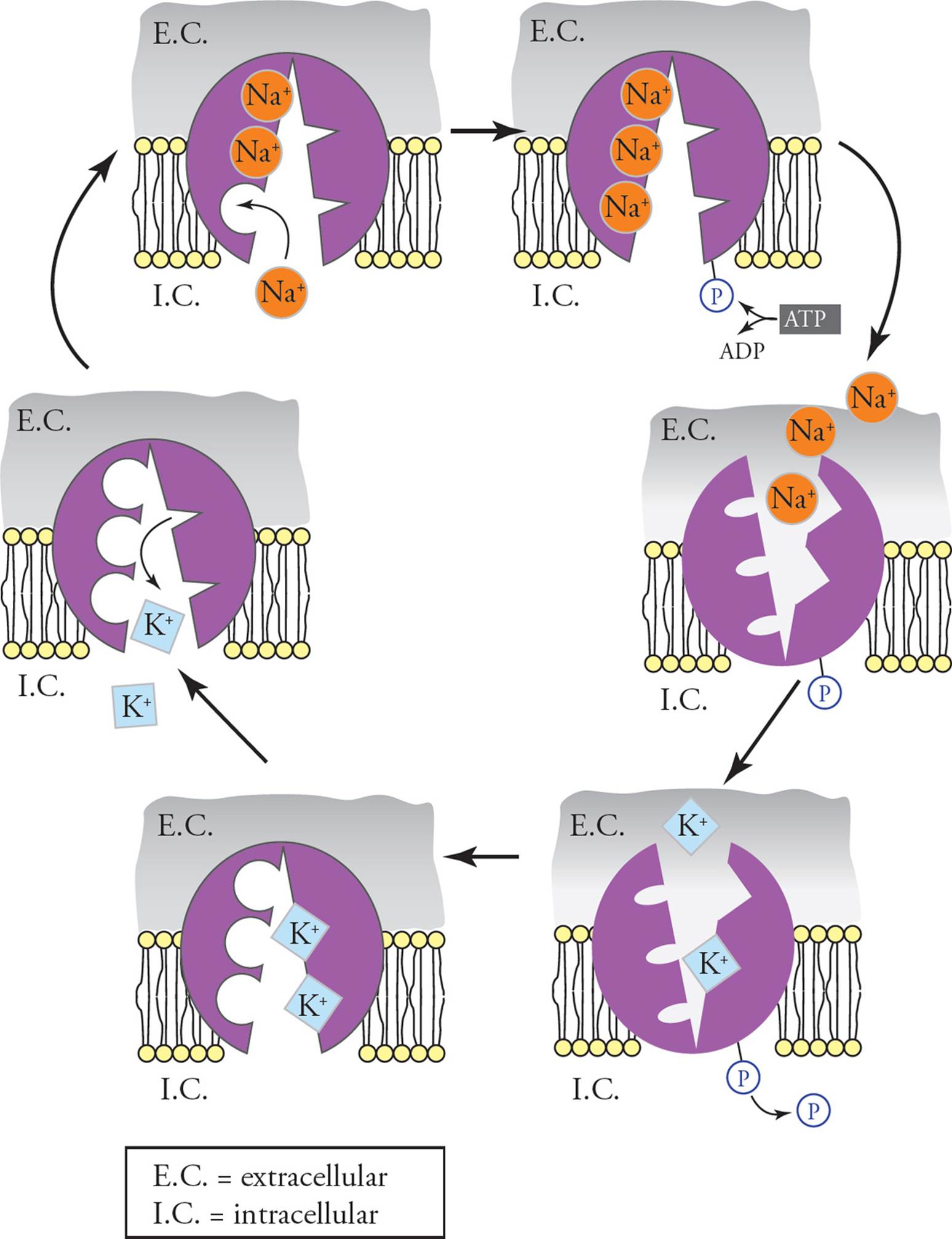
Figure 17 The Na+/K+ ATPase
How do we know exactly how the resting membrane potential is generated? For instance, how can we state with confidence that the electrogenicity of the Na+/K+ pump is far less important than the passive efflux of potassium in the generation of the RMP? The answer, given in the next two paragraphs, is not core MCAT material for memorization, but it is just the sort of thing that could show up in a passage.
The answers were determined using experiments. An artificial cell with no pumps and no channels in its membrane would have identical concentrations and charges inside and outside. An artificial cell with potassium leak channels but no active transporters would also obviously have no gradients across its membrane.
What about an artificial cell with Na+/K+ ATPase pumps and normal cellular concentrations of ATP and ADP + Pi but no potassium leak channels? Here is where experimentation was necessary. In this situation, the resting membrane potential is determined only by the electrogenicity of the Na+/K+ pump. The RMP in such a system turns out to be about –10 mV. [Why is it necessary to specify normal cellular concentrations of ATP and ADP + Pi? E.g., what would happen if there were much, much more ADP + Pi than ATP, as well as very high extracellular Na+ concentration and very high intracellular K+ concentration, in the artificial cell?38]
When K+ leak channels are added to the membrane (in addition to the Na+/K+ ATPase pumps and normal cellular concentrations of ATP and ADP + Pi), the RMP is measured at the normal cellular level, around −70 mV.
The following Table gives the concentrations of Na+, K+, and Cl– inside and outside the cell. (Know trends; don’t memorize numbers.) You already know why the Na+ and K+ concentrations are as they are. [Why is chloride so concentrated outside the cell?39] A useful mnemonic is to remember that life evolved in the ocean, which has very high concentrations of NaCl; hence the concentrations of Na+ and Cl– are high outside the cell and low inside.
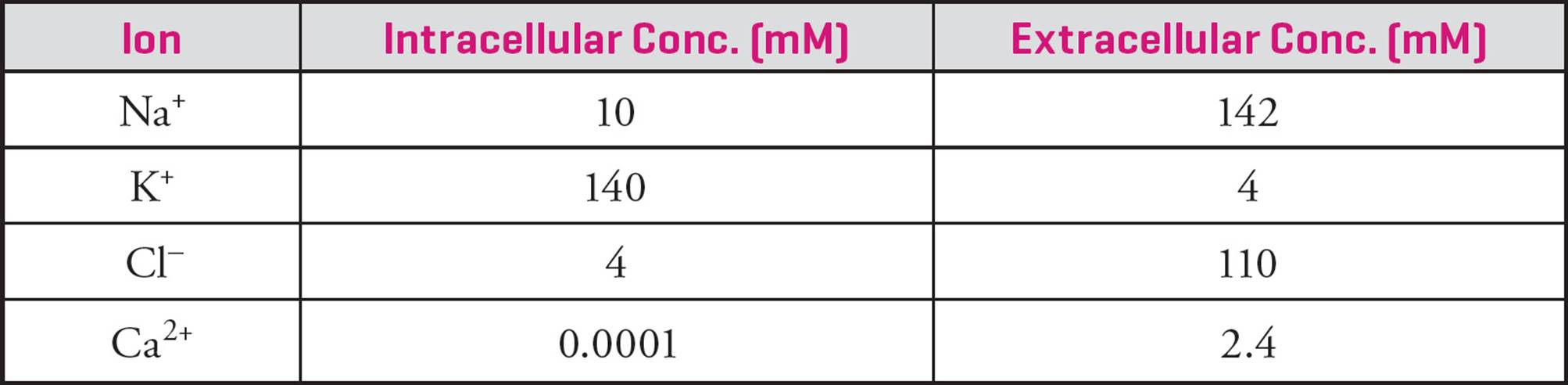
Table 3 Concentrations of Ions Inside/Outside Cell
Endocytosis and Exocytosis
Another mechanism used to transport material through the plasma membrane is within membrane-bound vesicles that fuse with the membrane (see Figure 18). Exocytosis is a process to transport material outside of the cell in which a vesicle in the cytoplasm fuses with the plasma membrane, and the contents of the vesicle are expelled into the extracellular space. The materials released are products secreted by the cell, such as hormones and digestive enzymes.
Endocytosis is the opposite of exocytosis: Generally, materials are taken into the cell by an invagination of a piece of the cell membrane to form a vesicle. Again, the cytoplasm is not allowed to mix with the extracellular environment. The new vesicle which is formed is called an endosome. There are three types of endocytosis:
1) Phagocytosis
2) Pinocytosis
3) Receptor-mediated endocytosis
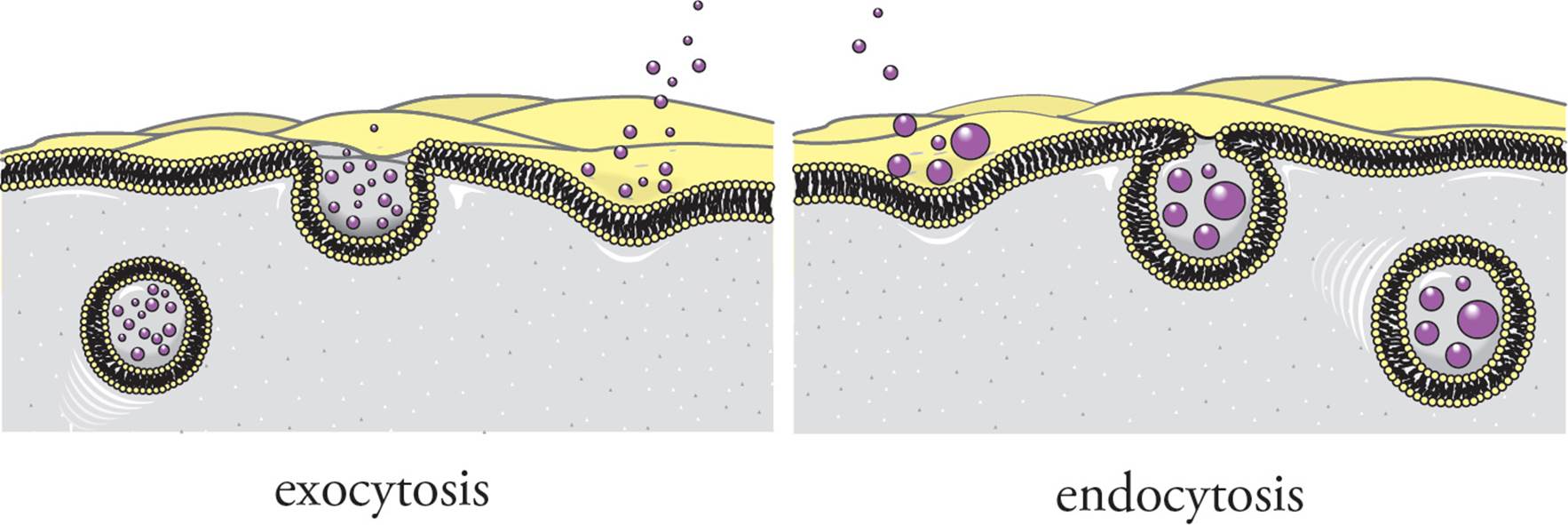
Figure 18 Endo- and Exocytosis
Phagocytosis means “cell eating.” It refers to the nonspecific uptake of large particulate matter into a phagocytic vesicle, which later merges with a lysosome. Thus, the phagocytosed material will be broken down. The prime example of phagocytic human cells are macrophages (“big eaters”) of the immune system, which engulf and destroy viruses and bacteria. (Note: This is not an invagination.)
Pinocytosis (cell drinking) is the nonspecific uptake of small molecules and extracellular fluid via invagination. Primitive eukaryotic cells obtain nutrition in this manner, but virtually all eukaryotic cells participate in pinocytosis.
Receptor-mediated endocytosis, on the other hand, is very specific. The site of endocytosis is marked by pits coated with the molecule clathrin (inside the cell) and with receptors that bind to a specific molecule (outside the cell). An important example is the uptake of cholesterol from the blood. Cholesterol is transported in the blood in large particles called lipoproteins. Cells obtain some of the cholesterol they require by receptor-mediated endocytosis of these lipoproteins. If they are not removed from the blood, cholesterol accumulates in the bloodstream, sticking to the inner walls of arteries. This results in atherosclerosis (a buildup of plaque on the walls of the arteries). [Does clathrin recognize and bind to lipoproteins?40] When the receptor-lipoprotein complex internalizes, it is taken into a vesicle that is termed an endosome. Lipoproteins are taken from the endosome to a lysosome where the cholesterol is released from the lipoprotein and the lipoprotein is degraded. The lipoprotein receptor is returned to the cell surface where it may again bind a lipoprotein. [How is receptor-mediated endocytosis similar to and different from active transport?41]
7.5 OTHER STRUCTURAL ELEMENTS OF THE CELL
Cell-Surface Receptors
Receptors form an important class of integral membrane proteins that transmit signals from the extracellular space into the cytoplasm. Each receptor binds a particular molecule in a highly specific lock-and-key interaction. The molecule that serves as the key for a given receptor is termed the ligand. The ligand is generally a hormone or a neurotransmitter. The binding of a ligand to its receptor on the extracellular surface of the plasma membrane triggers a response within the cell, a process termed signal transduction. Many cancers result from mutant cell-surface receptors which constitutively relay their signal to the cytoplasm, whether ligand is present or absent. For example, a growth factor exerts its effects by binding to a cell-surface receptor, and constitutive activity of a receptor for the growth factor causes uncontrolled growth of the cell. There are three main types of signal-transducing cell-surface receptors: ligand-gated ion channels, catalytic receptors, and G-protein-linked receptors.
Ligand-gated ion channels in the plasma membrane open an ion channel upon binding a particular neurotransmitter. An example is the ligand-gated sodium channel on the surface of the muscle cell at the neuromuscular junction. When the neurotransmitter acetylcholine binds to this receptor, the receptor undergoes a conformational change and becomes an open Na+ channel. The result is a massive influx of sodium down its electrochemical gradient, which depolarizes the muscle cell and causes it to contract.
Catalytic receptors have an enzymatic active site on the cytoplasmic side of the membrane. Enzyme activity is initiated by ligand binding at the extracellular surface. Generally, the catalytic role is that of a protein kinase, which is an enzyme that covalently attaches phosphate groups to proteins. Proteins can be modified with phosphate on the side chain hydroxyl of serine, threonine, or tyrosine. The insulin receptor is an example of a tyrosine kinase. Modification of proteins with phosphates regulates their activity.
A G-protein-linked receptor does not directly transduce its signal, but transmits it into the cell with the aid of a second messenger. This is a chemical signal that relays instructions from the cell surface to enzymes in the cytoplasm. The most important second messenger is cyclic AMP (cAMP). It is known as a “universal hunger signal” because it is the second messenger of the hormones epinephrine and glucagon, which cause energy mobilization (glycogen and fat breakdown). Second messengers such as cAMP allow a much greater signal than receptor alone produces (see Figure 19). An epinephrine molecule activates one G-protein-linked receptor which activates many G-proteins, each G-protein activates many adenylyl cyclase enzymes, each adenylyl cyclase makes lots of cAMP from ATP, each cAMP activates many cAMP-dPK, and each cAMP-dPK phosphorylates many enzymes. Some of these enzymes will be activated, and others inactivated by phosphorylation, with the end result that the entire cell harmoniously works toward the same goal: energy mobilization.
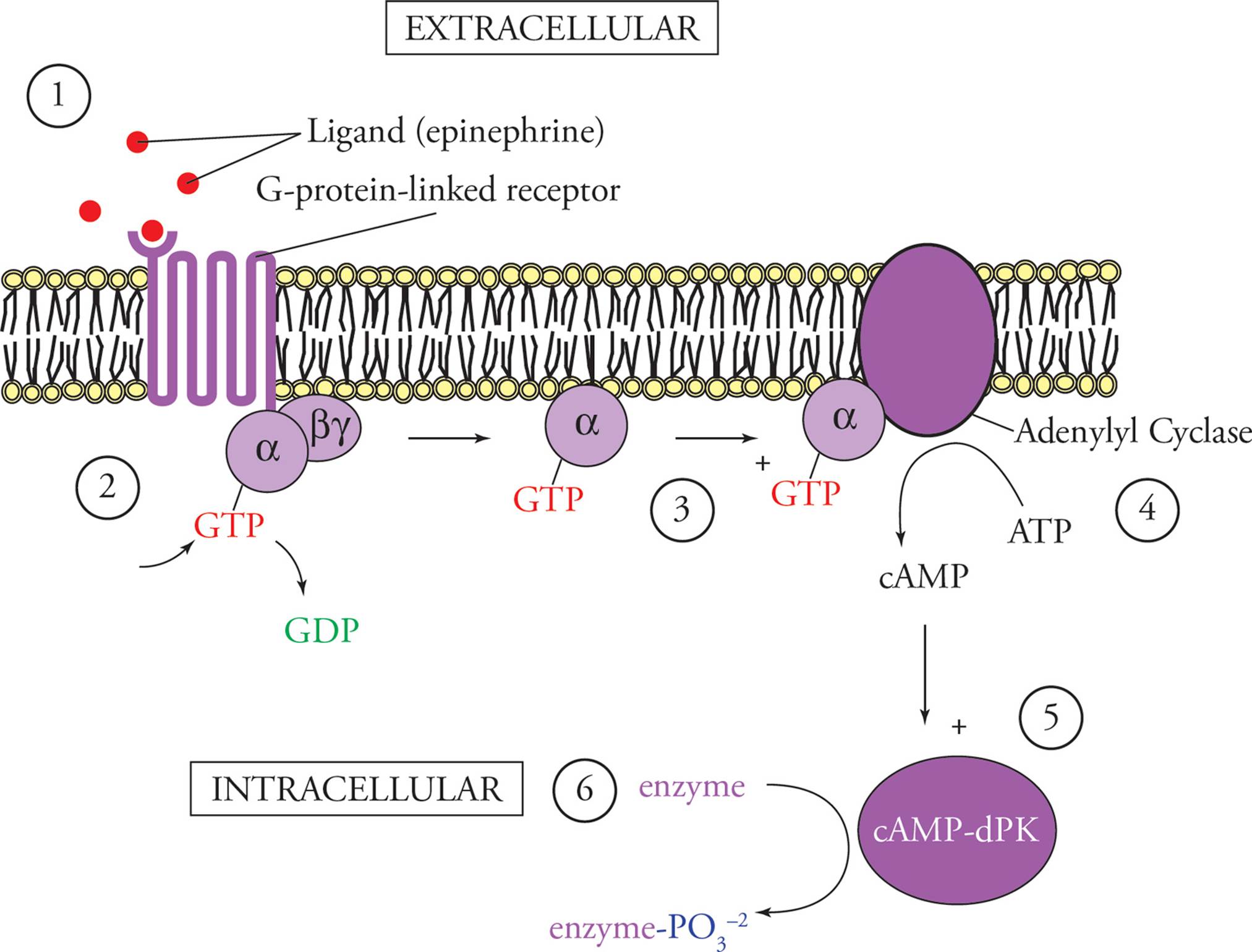
Figure 19 G-Protein Mediated Signal Transduction Stimulated by Epinephrine
1) Epinephrine arrives at the cell surface and binds to a specific G-protein-linked receptor.
2) The cytoplasmic portion of the receptor activates G-proteins, causing GDP to dissociate and GTP to bind in its place.
3) The activated G-proteins diffuse through the membrane and activate adenylyl cyclase.
4) Adenylyl cyclase makes cAMP from ATP.
5) cAMP activates cAMP-dependent protein kinases (cAMP-dPK) in the cytoplasm.
6) cAMP-dPK phosphorylates certain enzymes, with the end result being mobilization of energy. For example, enzymes necessary for glycogen breakdown will be activated, while enzymes necessary for glycogen synthesis will be inactivated, by cAMP-dPK phosphorylation.
There are different types of G-protein-linked receptors. The one depicted above is a stimulatory one. Its G-protein would be denoted Gs. Inhibitory G-protein-linked receptors activate inhibitory G-proteins (Gi) which serve to inactivate adenylyl cyclase instead of activating it. In this way different hormones can modulate each other’s effects.
There are also G-protein-linked receptors which have nothing to do with cAMP. Instead, their G-proteins activate an enzyme called phospholipase C, initiating a different second messenger cascade, which results in an increase in cytoplasmic Ca2+ levels. The common theme shared by all G-protein-based signal transduction systems is their reliance on a G-protein, which is a signaling molecule that binds GTP. You should understand these key notions: cAMP as a second messenger, signal transduction, and signal amplification. The remaining details are not important for the MCAT; read for concepts, not memory.
The Cytoskeleton
The animal cell cytoskeleton provides the structural support supplied by the cell wall in bacteria, plants, and fungi. It also allows movement of the cell and its appendages (cilia and flagella) and transport of substances within the cell. Animal cells have an internal cytoskeleton composed of three types of proteins: microtubules, intermediate filaments, and microfilaments (see Figure 20). Microtubules are the thickest, microfilaments the thinnest. All three are composed of noncovalently polymerized proteins; in other words, they are a massive example of quaternary protein structure.
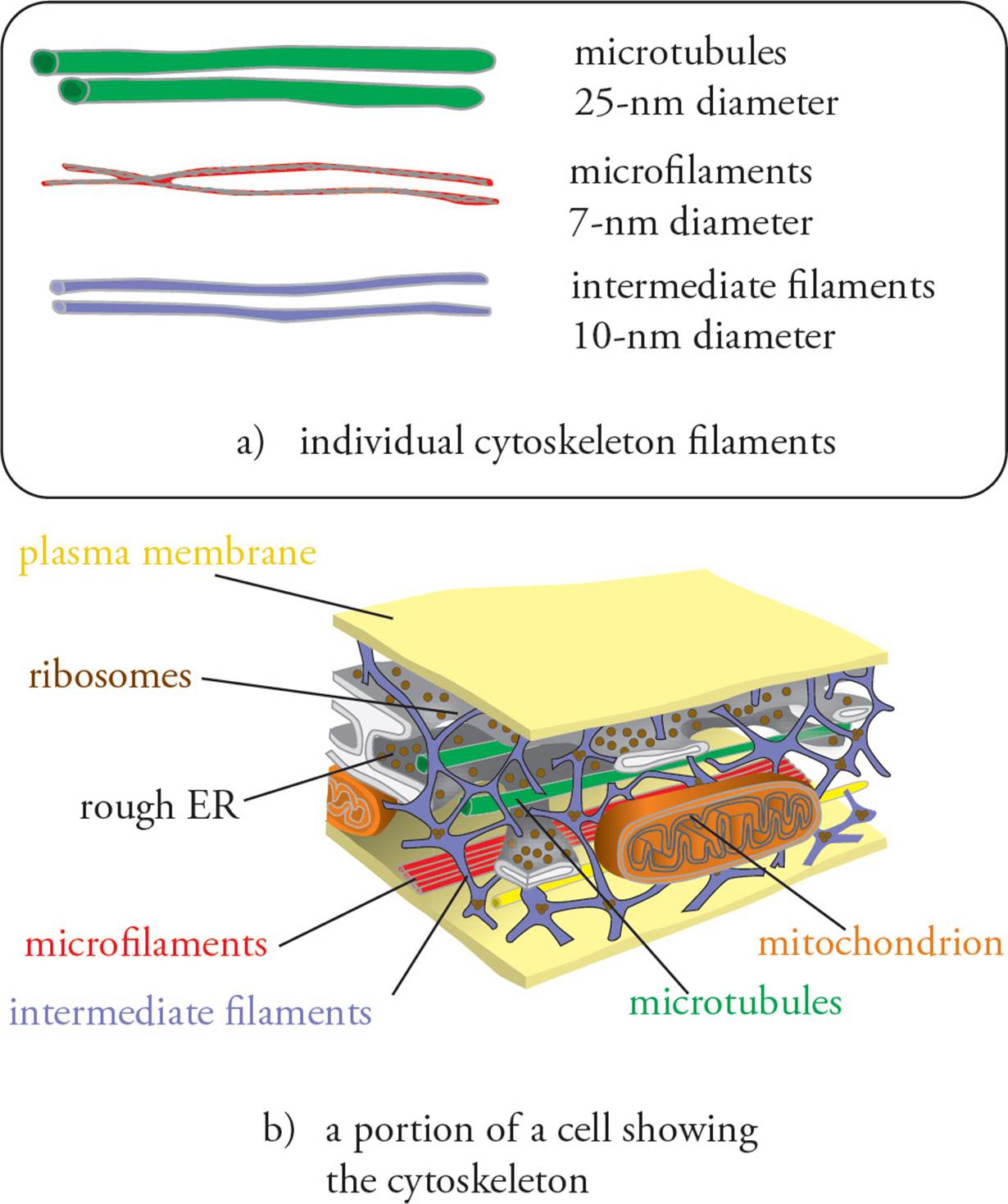
Figure 20 Cytoskeleton
Microtubules
The microtubule is a hollow rod composed of two globular proteins: α-tubulin and β-tubulin, polymerized noncovalently. First, α-tubulin and β-tubulin form an αβ-tubulin dimer. Then many dimers stick to each other noncovalently to form a sheet, which rolls into a tube. Once formed, the microtubule can elongate by adding αβ-tubulin dimers to one end. The other end cannot elongate, because it is anchored to the microtubule organizing center(MTOC), located near the nucleus. Microtubules are dynamic and can get longer or shorter by adding or removing tubulin monomers from the end.
Within the MTOC is a pair of centrioles (see Figure 21). Each centriole is composed of a ring of nine microtubule triplets. When cell division occurs, the centrioles duplicate themselves, and then one pair moves to each end of the cell. During mitosis, microtubules radiating out from the centrioles attach to the replicated chromosomes and pull them apart so that one copy of each chromosome (one chromatid) moves to each end of the cell. The resulting daughter cells each get a full copy of the genome plus a centriole pair. The microtubules that radiate out from the centrioles during mitosis are called the aster, because they are star-shaped. The microtubules connecting the chromosomes to the aster are polar fibers. The whole assembly is called the mitotic spindle. The centromere of each chromosome contains a kinetochore which is attached to the spindle by tiny microtubules called kinetochore fibers. Refer to the Figure in the upcoming section on mitosis.
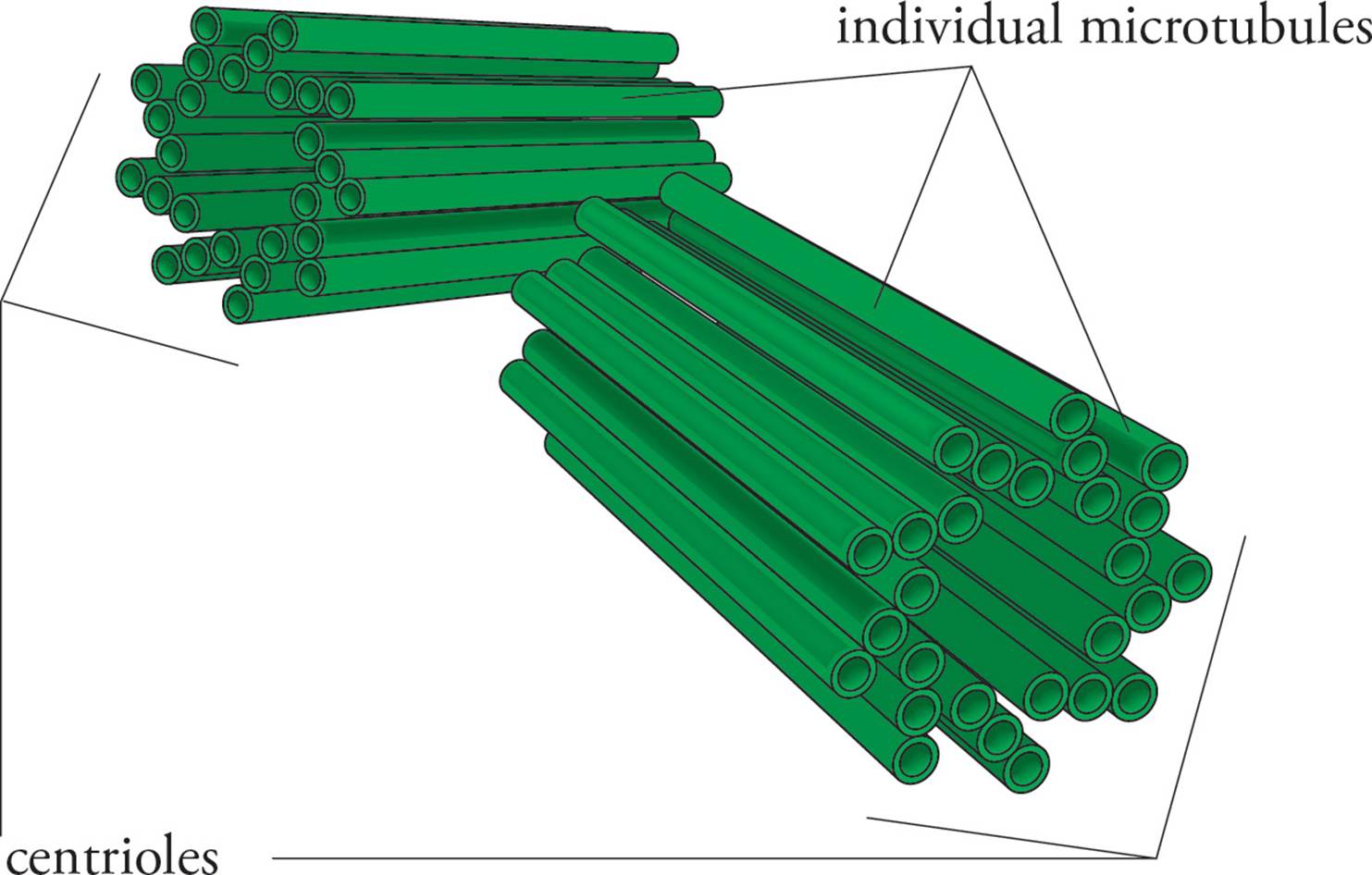
Figure 21 A Pair of Centrioles
In mitosis, the MTOC is essential, but the centrioles are not. There are two major pieces of evidence for this: 1) Plant cells lack centrioles but still undergo mitosis; 2) Experimenters have succeeded in removing the centrioles from animal cells, and the cells were still able to undergo mitosis.
Microtubules also mediate transport of substances within the cell. In nerve cells, materials are transported from the cell body to the axon terminus on a microtubule railroad. The transport process is driven by proteins that hydrolyze ATP and act as molecular motors along the microtubule.
Eukaryotic Cilia and Flagella
Cilia are small “hairs” on the cell surface which move fluids past the cell surface. For example, cilia on lining cells of the human respiratory tract continually sweep mucus toward the mouth in a mechanism termed the mucociliary escalator. A flagellum is a large “tail” which moves the cell by wiggling. The only human cell which has a flagellum is the __.42 Cilia are small and flagella are long, but they have the same structure, with a “9 + 2” arrangement of microtubules (see Figure 22). Nine pairs of microtubules form a ring around two lone microtubules in the center. Each microtubule is bound to its neighbor by a contractile protein called dynein which causes movement of the filaments past one another. The cilium or flagellum is anchored to the plasma membrane by a basal body, which has the same structure as a centriole (a ring of nine triplets of microtubules). Remember that the prokaryotic flagellum is different in structure, and its motion is driven by a different mechanism.
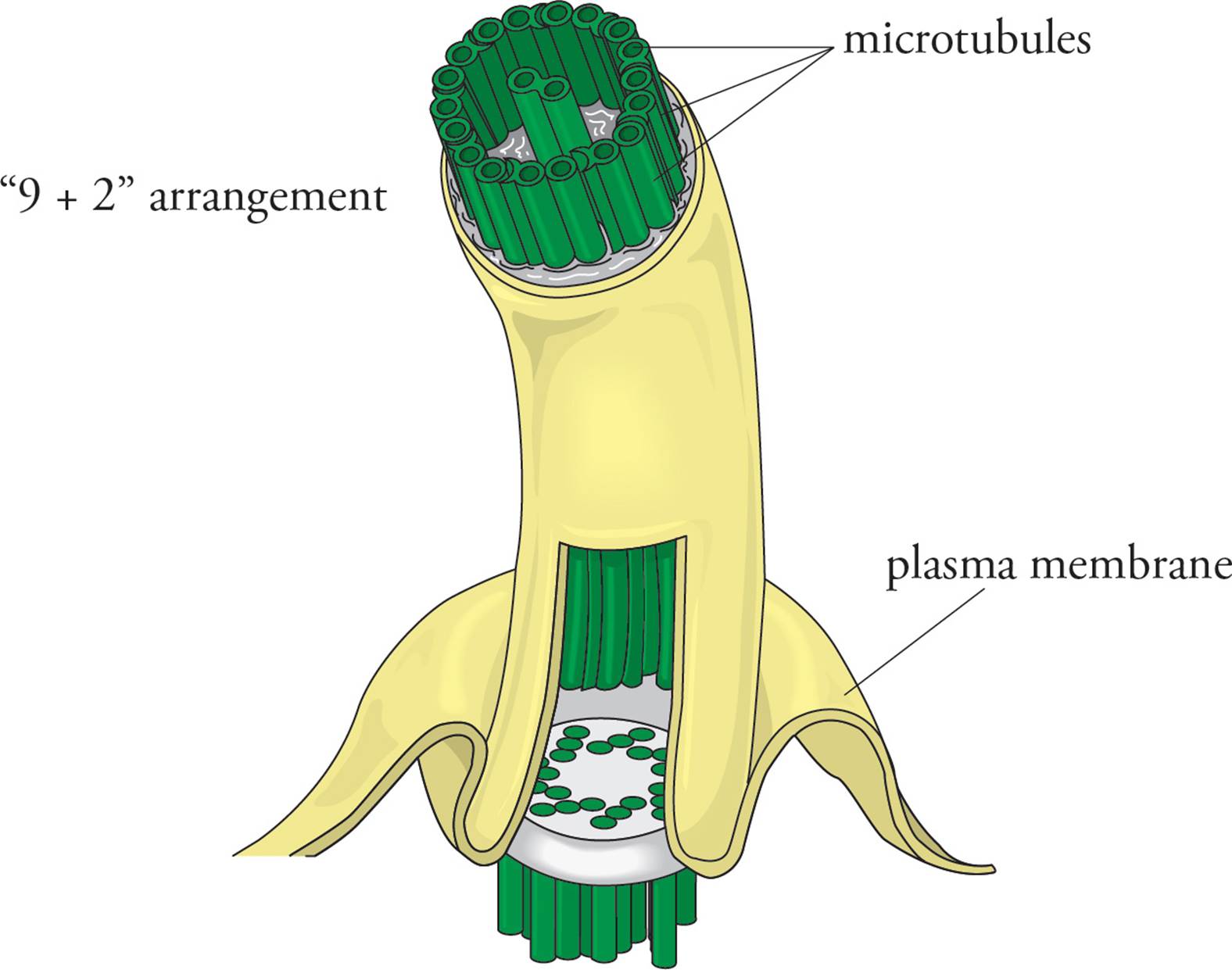
Figure 22 The Base of a Cilium or Flagellum
Microfilaments
Microfilaments are rods formed in the cytoplasm from polymerization of the globular protein actin. Actin monomers form a chain, and then two chains wrap around each other to form an actin filament. Microfilaments are dynamic and are responsible for gross movements of the entire cell, such as pinching the dividing parent cell into two daughters during cell division, and amoeboid movement. Amoeboid movement involves changes in the cytoplasmic structure which cause cytoplasm and the rest of the cell to flow in one direction.
Intermediate Filaments
Intermediate filaments are named for their thickness, which is between that of microtubules and microfilaments. Unlike microtubules and microfilaments, intermediate filaments are heterogeneous, composed of a wide range of polypeptides. Another difference is that intermediate filaments are more permanent, whereas microfilaments and microtubules are often disassembled and reassembled as needed by the cell. Intermediate filaments appear to be involved in providing strong cell structure, such as in resisting mechanical stress.
Cell Adhesion and Cell Junctions
In some tissues, cells are tightly bound to each other. For example, the intestinal wall is lined with a type of tissue called epithelium.43 The layer of epithelial cells in the gut forms a tight seal, preventing items from moving freely between the intestinal lumen and the body; this is accomplished by tight junctions.
Epithelial cells in the skin are held together tightly but do not form a complete seal; this is accomplished by desmosomes. Some specialized cell types, such as heart muscle cells, are connected by holes called gap junctions that allow ions to flow back and forth between them. We discuss each of the above structures in the following paragraphs (see Figure 23).
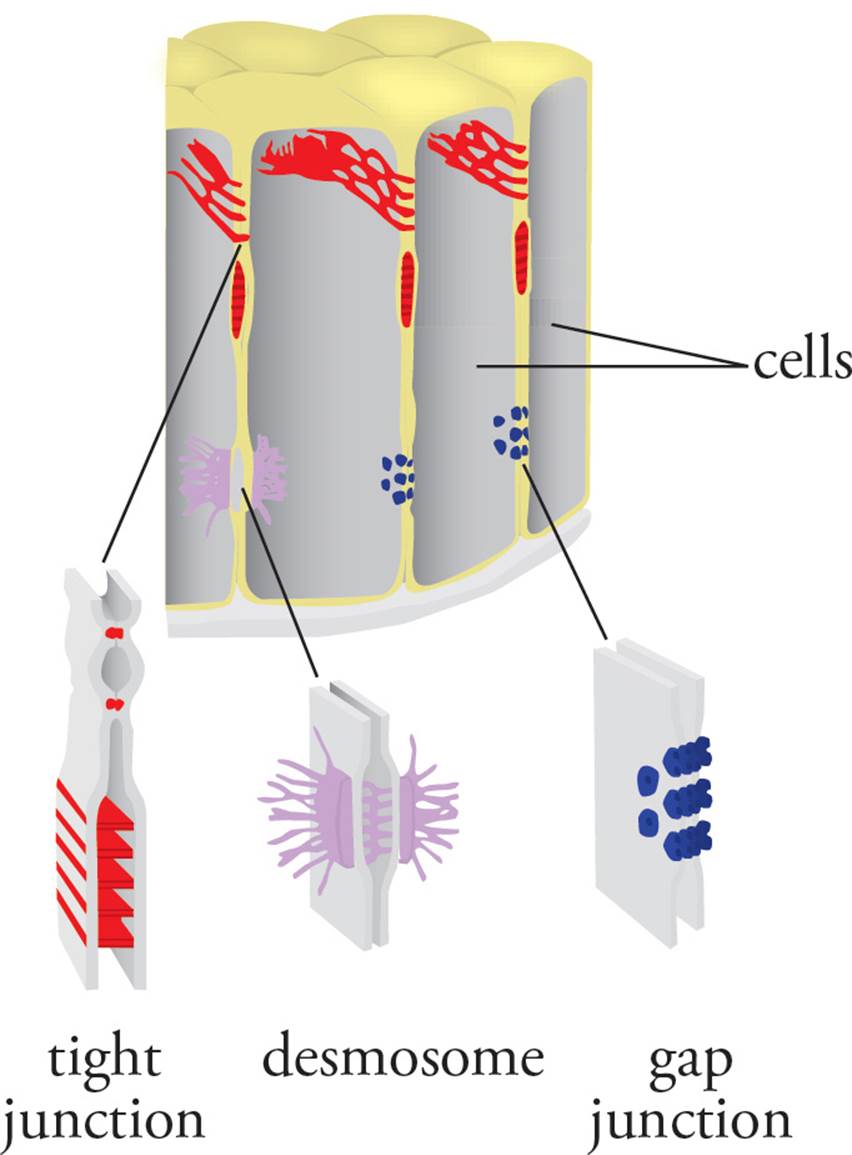
Figure 23 Cell Junctions
Tight junctions are also termed occluding junctions because they do not just join cells at one point, but form a seal between the membranes of adjacent cells that blocks the flow of molecules across the entire cell layer. They are not spots where cells are stuck together, but rather bands running all the way around the cells. Intestinal epithelial cells are involved in the active transport of glucose and other molecules from one side of epithelium to the other. A tight seal between these cells is required to prevent the two compartments from mixing. Tight junctions also block the flow of molecules within the plane of the plasma membrane. For example, the surface of the plasma membrane facing the intestinal lumen, termed the apical surface, has different membrane proteins than the plasma membrane on the other side of the cell facing the tissues beneath, called the basolateral surface. [Will a transmembrane protein inserted into the apical surface of an intestinal epithelial cell diffuse in the plane of the plasma membrane to reach the basolateral surface of the cell?44]
Desmosomes do not form a seal, but merely hold cells together; they are also known as spot desmosomes because they are concise points, not bands all the way around the cell. The desmosome is composed of fibers that span the plasma membranes of two cells. Inside each cell, the desmosome is anchored to the plasma membrane by a plaque formed by the protein keratin. Intermediate filaments of the cytoplasm attach to the inside of the desmosome. Desmosomes do not freely diffuse in the plane of the plasma membrane, as suggested by the fluid mosaic model, because they are anchored in place by intermediate filaments of the cytoskeleton. As you can see, the fluid mosaic model is an idealization describing the plasma membrane in pure form. In the real cell membrane, things are highly organized.
Gap junctions form pore-like connections between adjacent cells, allowing the two cells’ cytoplasms to mix. The connection is large enough to permit the exchange of solutes such as ions, amino acids, and carbohydrates, but not polypeptides and organelles. Gap junctions in smooth muscle and cardiac muscle allow the membrane depolarization of an action potential to pass directly from one cell to another.
7.6 THE CELL CYCLE AND MITOSIS
Our cells must reproduce themselves in order to replace lost or damaged cells and so that tissues can grow. Cells reproduce themselves by first doubling everything in the cytoplasm and the genome and then splitting in half. Some cells continually go through a cycle of growth and division, which is traditionally discussed in four phases (see Figure 24). S (synthesis) phase is when the cell actively replicates its genome, as described in Chapter 3. M phaseincludes mitosis and cytokinesis. Mitosis is the partitioning of cellular components (genes, organelles, etc.) into two halves. Cytokinesis is the physical process of cell division. Between M phase and S phase, there are two “gap” phases, G1 and G2. The gap phases plus S phase together form the part of the cell cycle between divisions, known as interphase.
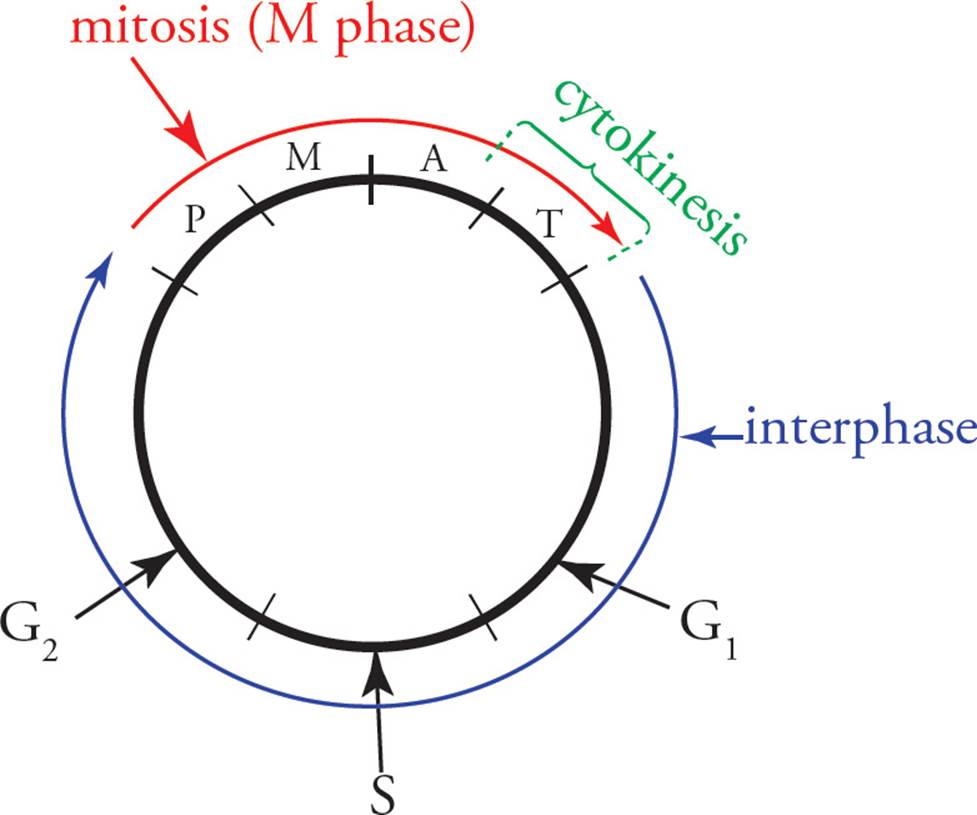
Figure 24 The Cell Cycle
The cell spends most of its time in interphase, busily metabolizing and synthesizing materials. Some cells are permanently stuck in interphase (G0). In fact, the more specialized a cell becomes, the less likely it is to remain capable of reproducing itself. Examples are neurons, blood cells, and cells on the surface of the skin. They must be replenished by reproduction of less specialized precursor cells called stem cells. All the blood cells, for example, are derived from a single type of stem cell found in the bone marrow.
During interphase, the genome is spread out in a form that is not visible with a light microscope without special stains, and DNA is accessible to the enzymes of replication. By the end of S phase, the nucleus contains two complete copies of the genome. The cell now has twice the normal amount of DNA.
Mitosis is divided into four phases: prophase, metaphase, anaphase, and telophase.45 The first sign of prophase is that the genome becomes visible upon condensing into densely-packed chromosomes, instead of diffuse chromatin. [Why do the chromosomes condense?46] Observing a human cell under the light microscope at the beginning of prophase, one can see 46 differently-shaped chromosomes. Upon closer observation, one notes that each chromosome actually consists of two identical particles joined at a centromere. These two particles are the two copies of a chromosome, known as sister chromatids. When mitosis is complete, each new daughter cell will have 46 chromosomes, each consisting of a single chromatid, separated from its sister. Spending a little more time staring at the nucleus, you might notice that the jumble of 46 chromatid pairs actually consists of 23 homologous pairs of identical-appearing sister chromatid pairs (23 pairs of pairs). Homologous chromosomes are different copies of the same chromosome, one from your mother and the other from your father. (Also refer to Chapter 6.) To repeat:
Sister chromatids are identical copies of a chromosome, attached to each other at the centromere. Homologous chromosomes are equivalent but nonidentical and do not come anywhere near each other during mitosis.
Other important events occur during prophase. The nucleolus disappears, the spindle and kinetochore fibers appear, and the centriole pairs begin to move to opposite ends of the cell. So now the cell has two MTOCs, called asters(stars) because of the star-like appearance of microtubules radiating out. Also at the end of prophase, the nuclear envelope converts itself into many tiny vesicles.47
Metaphase is simple: All the chromosomes line up at the center of the cell, forming the metaphase plate. The chromosomes line up in the center of the cell because the kinetochore of each sister chromatid is attached to spindle fibers that attach to MTOC at opposite ends of the cell. So each member of a pair of chromatids is pulled toward the opposite pole of the cell.
During anaphase, the spindle fibers shorten, and the centromeres of each sister chromatid pair are pulled apart. The cell elongates, and cytokinesis begins with the formation of a cleavage furrow, which is accomplished by __.48
In telophase (telos is Greek for “end”), a nuclear membrane forms around the bunch of chromosomes at each end of the cell, the chromosomes decondense, and a nucleolus becomes visible within each new daughter nucleus. Each daughter nucleus has 2n chromosomes. Cytokinesis is complete, and the cell is split in two (see Figure 25).
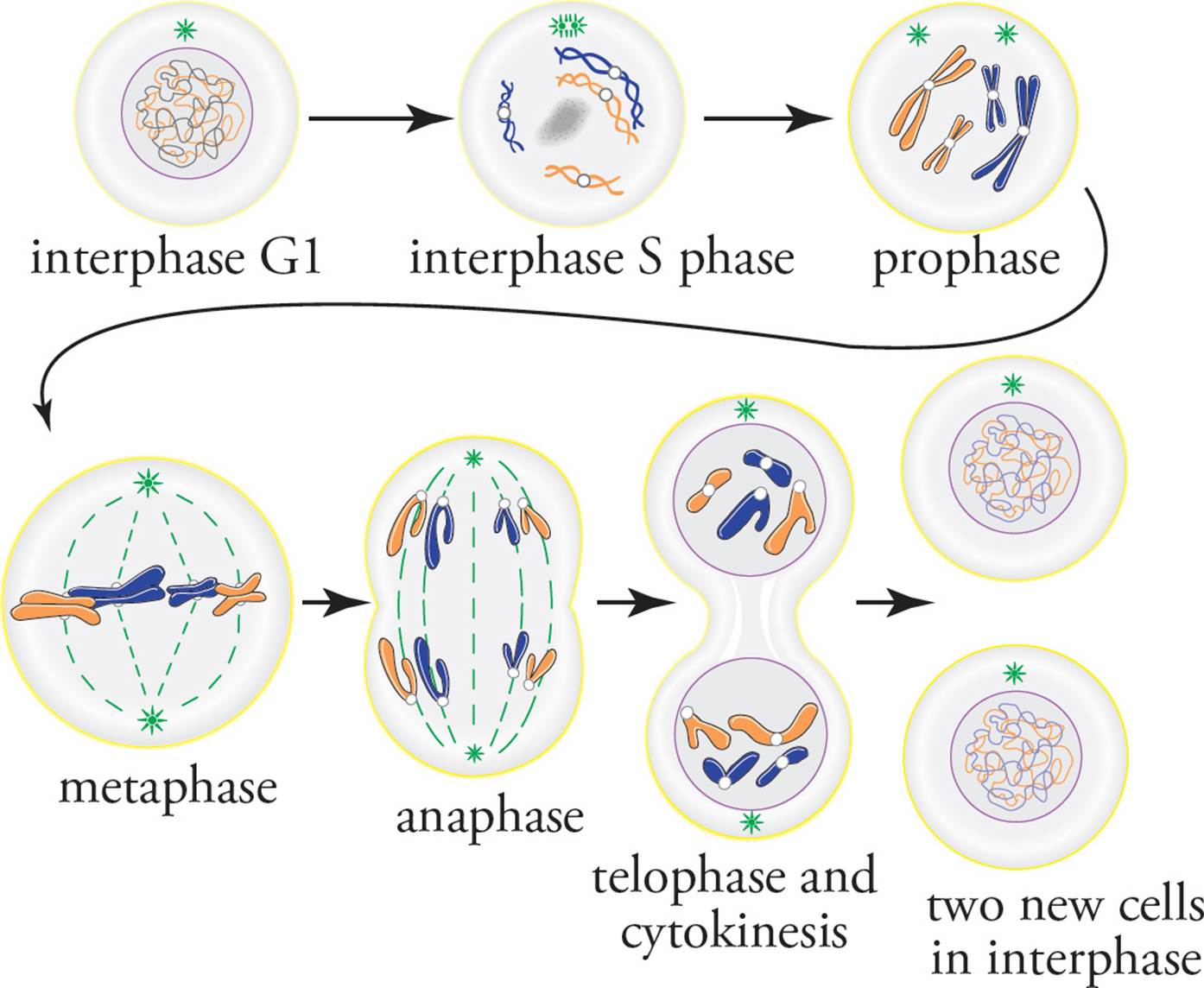
Figure 25 The Phases of Mitosis
The karyotype is a display of an organism’s genome (see Figure 26). A cell is frozen during metaphase, its chromosomes are stained, and a photograph is taken. The micrograph is enlarged, and each chromosome is cut out of the picture with an artist’s blade. Then all homologues are paired, and the entire genome is examined for abnormalities.
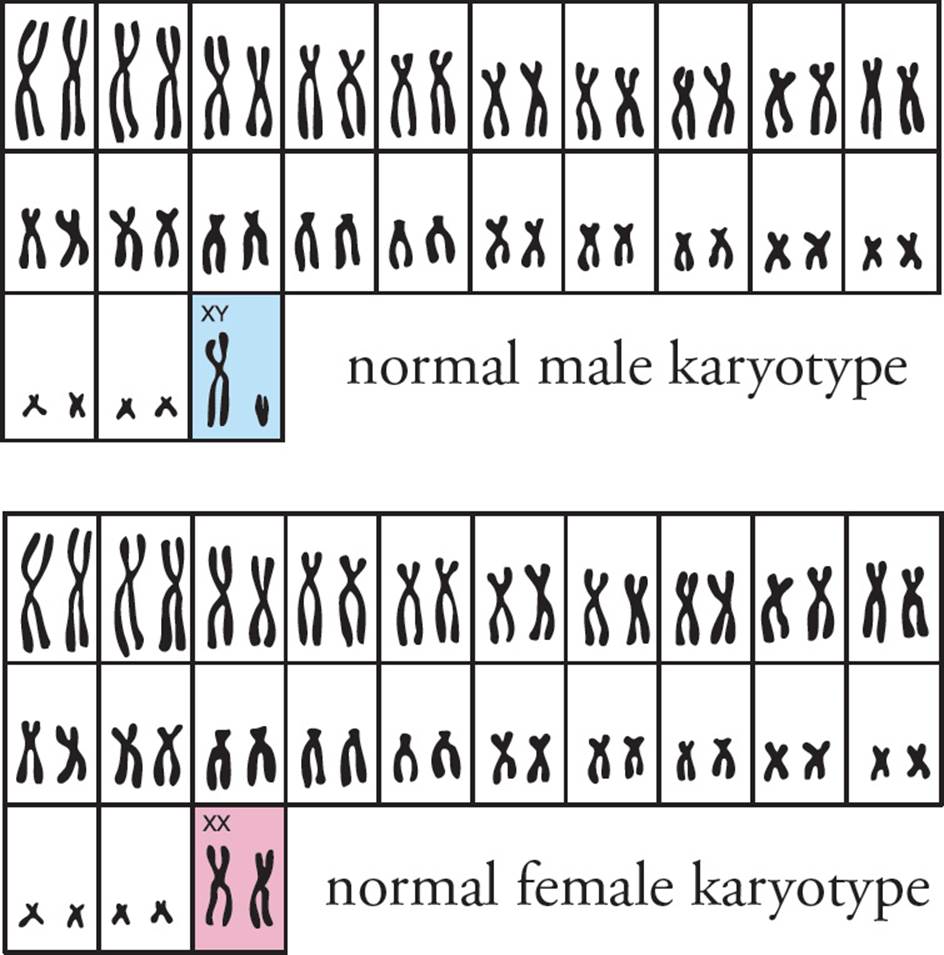
Figure 26 A Genetic Karyotype
• Eukaryotic chromosomes generally have only one of which of the following?49
A) Reading frame
B) Origins of replication
C) Promoter
D) Centromere
7.7 CANCER, ONCOGENES, AND TUMOR SUPPRESSORS
Inappropriate cell division (i.e., cells that have lost control of the cell cycle) can have disastrous consequences. A mutation in a protein that is normally involved in regulating progression through the cell cycle can result in unregulated cell division and cancer. Cancer means “crab,” as in the zodiac sign. The name derives from the observation that malignant tumors grow into the surrounding tissue, embedding themselves like clawed crabs.
• In normal eukaryotic cells, mitosis will not begin until the entire genome is replicated. If this inhibition is removed so that mitosis begins during S-phase, which one of the following would occur?50
A) The cells would grow more quickly.
B) The genome would become fragmented and incomplete.
C) The cells would display unregulated, cancerous growth.
D) The genome would be temporarily incomplete in each daughter cell, but DNA repair will fill in the missing gaps.
Cancers can present as malignant solid tumors or in a more diffuse cellular state, such as leukemia, a cancer occurring in the bone marrow where improper leukocytes are formed and circulated.
Mutated genes that induce cancer are termed oncogenes (“onco-” is a prefix denoting cancer). Normally, these genes are required for proper growth of the cell and regulation of the cell cycle. Oncogenes, then, are genes that can convert normal cells into cancerous cells. Sometimes these are abnormal versions of standard cellular growth genes. Sometimes the genes enter the cell because of a viral infection. In fact, the first identified oncogene, labeled src, was isolated from a retrovirus found in chickens; the oncogene contributes to sarcomas, cancers of the bone, cartilage, adipose, muscular, vascular or hematopoietic tissues. [Teratomas are tumors with formed tissues from multiple germ layers. What steps might lead to their formation?51]
Protooncogenes are the normal versions of the genes that allow for regular growth patterns, but can be converted into oncogenes under the right circumstances. Conversion may be due to mutation or because of exposure to a mutagen. Ultraviolet radiation (such as sunlight or light from tanning booths) and various chemicals (such as benzene) are both examples of common mutagens.
Tumor suppressor genes produce proteins that are the inherent defense system to prevent the conversion of cells into cancer cells. The two primary means of cancer prevention are to (a) detect damage to the genome and halt cell growth and division until the damage can be repaired, or (b) to trigger programmed cell death if the damage is too severe to be repaired. p53 is an example of a product of a common tumor suppressor gene. Though normally at low levels in cells, its production is scaled up when genetic damage or oncogene activity is detected, and if sufficient repair is not possible, p53 will cause the cell to die in a process referred to as apoptosis.
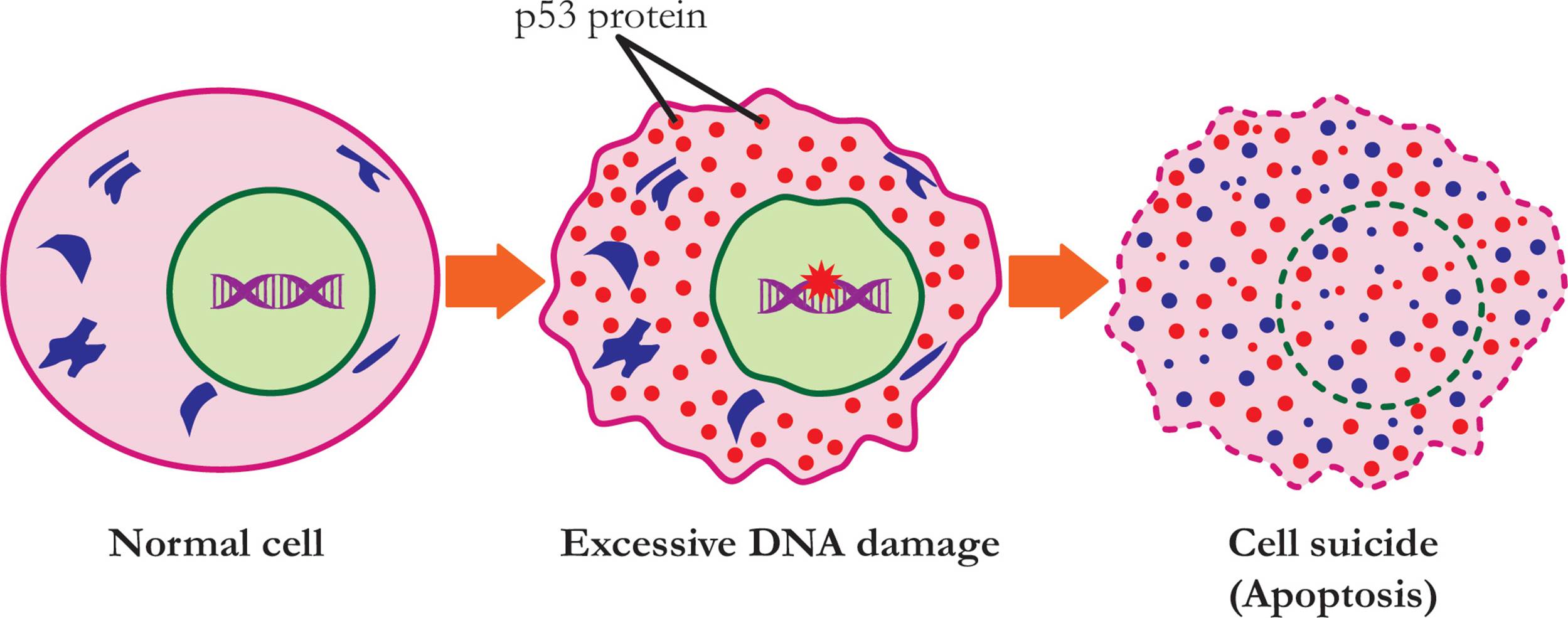
Figure 27 Representation of p53 triggering apoptosis
Apoptosis
Apoptosis, or programmed cell death, allows a cell to shrink and die while simultaneously minimizing damage to neighboring cells and limiting the exposure of other cells to its cytosolic contents. The death of a cell is triggered by a stressor which may be external (such as nitric oxide, a toxin, or cytokines) or internal (such as when the level of the p53 tumor suppressor protein reaches a critical level).
The process of apoptosis begins with the shrinking of the cell and the disassembly of the cytoskeleton. While the cellular infrastructure is taken apart, the nuclear envelope breaks down and the genome is broken into pieces. A different profile of cell surface proteins emerges, thus signaling various phagocytic cells, including macrophages, to finish deconstructing and clearing away the dead cell.
A family of proteases, referred to as caspases, is responsible for carrying out the events of apoptosis. They have a cysteine in their active site and they cleave their target proteins at aspartic acid sites, hence their name (c-asp-ases). Caspases, like all potentially damaging enzymes, are produced in their inactive form as procaspases. Twelve different caspases have been identified in humans, and they are generally grouped into two categories, initiators and effectors. Initiator caspases respond to extra- or intracellular death signals by clustering together; this clustering allows them to activate each other. The activation of the initiators leads to the activation of the effector caspases in a cascade of activation. Effector caspases then cleave a variety of cellular proteins to trigger apoptosis.
Figure 28 Caspase Activation and Cascade
Oxidative Stress
Oxidative stress occurs when the level of production of reactive oxygen species outstrips the cell’s ability to detoxify them. This can include increased levels of peroxides or free oxygen, which then generates radicals. While reactive oxygen species are normally produced as part of metabolism, at high untreated levels they can damage DNA, cellular proteins, and even lipid bilayers. As such, oxidative stress is linked to cancer, since the damage it can cause sets up conditions in the cell to allow oncogenes to become active and cell growth to be impacted.
Though the potential for damage exists, creating oxidative stress is also a component of the immune system. Activated phagocytes may produce nitric oxide or superoxide (O2–) in order to broadly kill pathogens. The effect on foreign cells needs to be balanced against the damage being caused to host tissues.
Regenerative Capacity
The ability to restore damaged tissues is an important survival strategy, and depends on how stem cells are maintained in the body of an organism. Planarians (simple flatworms) have collections of stem cells throughout their bodies that can migrate to areas of damage and rebuild necessary tissues. Newts or lizards that lose a limb have cells that dedifferentiate and then redifferentiate to grow back all the components of that limb. Within mammals, however, the regenerative capacity is considerably more limited. The pluripotent stem cells within the bone marrow are constantly regenerating the cellular components of the blood and immune system. Hepatocytes in the liver divide to produce more cells, but this process is much slower; these cells are described as being unipotent. Some cells, like neurons, do not regenerate, though inducing them to do so is an active area of research so as to better address injuries and damage caused by neurological degrading illnesses. [The benefits of having regenerative capacity are clear, but if it does not proceed properly, what could be the outcome?52]
Senescence
Senescence describes the process of biological aging which occurs at both the cellular and the organismal level. For eukaryotic cells, the length of the telomeres on the ends of chromosomes are a measure of cellular age; the longer the telomeres, the younger the cell. These sequences are meant to be maintained by the enzyme telomerase (Chapter 5). Research is being pursued as to whether the biological age of a cell can be reset if telomerases are manipulated to maintain the length of the telomeres. Additionally, as cells age they become prone to apoptosis. Though stressors can induce apoptosis earlier than expected, this mechanism of programmed cell death is also how organisms destroy and disassemble cells that need to be removed due to age.
The cumulative effects of cellular senescence lead to the aging of the entire organism. The functioning of organs is affected to the point where the body stops working and death occurs. These effects can be hastened based on environmental exposures and behavioral factors, but even without additional stressors, senescence is ineviTable for organisms.
Chapter 7 Summary
• For the MCAT, you should know the structures and functions of the following key eukaryotic organelles: nucleus, mitochondria, ribosomes, rough ER, smooth ER, Golgi apparatus, lysosomes, and peroxisomes.
• The rough ER is the site of translation of proteins to be either secreted from the cell, inserted into the membrane, or targeted to the lysosomes, ER, or Golgi apparatus.
• Signal sequences are specific amino acid sequences that direct proteins in translation to the rough ER and the secretory pathway (rough ER → Golgi apparatus → final location).
• Post-translational modification can occur in the rough ER or in the Golgi apparatus.
• All cellular membranes are composed of lipid bilayers with distinct hydrophobic and hydrophilic regions. The membranes act as selective barriers that regulate which molecules can cross.
• An electrolyte is a solute that produces free ions in solution. Strong electrolytes produce more ions in solution than weak electrolytes.
• The van’t Hoff (or ionizability) factor, i, tells us how many ions one unit of a substance will produce in solution.
• Colligative properties depend on the number of particles in solution. Colligative properties include vapor pressure depression, boiling point elevation, freezing point depression, and osmotic pressure.
• Molecules naturally want to move from regions of higher concentration to regions of lower concentration (with respect to that particular molecule). Diffusion is the movement of particles down their concentration gradient, and osmosis is the movement of water down its concentration gradient.
• Hydrophobic molecules (e.g., O2, CO2, and steroids) cross the membrane by simple diffusion, while hydrophilic, polar molecules (e.g., ions, glucose, and water) must cross the membrane with the help of a special membrane protein (a channel or a carrier). This is called facilitated diffusion.
• Active transport uses energy to move molecules against their concentration gradients (from low concentration areas to higher concentration areas). Primary active transport uses ATP directly, while secondary active transport relies on gradients previously established by a primary active transporter.
• The Na+/K+ ATPase is a primary active transporter that moves three Na+ ions out of the cell for every two K+ ions it moves into the cell. This helps establish the resting membrane potential of the cell, helps maintain osmotic balance in the cell, and sets up a Na+ gradient that can be used for secondary active transport.
• G-proteins help transduce signals from extracellular ligands across the membrane. They change the level of cAMP or calcium (second messengers) in the cell, which changes the metabolic enzyme pathways active in the cell.
• Microtubules form centrioles, cilia, and eukaryotic flagella, while microfilaments participate in contractile activity.
• Tight junctions help form a seal between cells so that the flow of molecules across the entire cell layer is regulated. Desmosomes form general adhesions between cells. Gap junctions form connections between cells that allow the flow of cytoplasm from cell to cell.
• During the cell cycle, DNA replication occurs during the S-phase of interphase, and cell division occurs during mitosis (M-phase).
• Mitosis is comprised of four major phases (prophase, metaphase, anaphase, and telophase) and results in two daughter cells that are identical to each other and identical to the original parent cell.
• Protooncogenes can be mutated to form oncogenes, which can lead to cancer. Tumor suppressor genes can prevent cancer by halting the cell cycle until damaged DNA is repaired or by inducing apoptosis (cell suicide).
• Apoptosis is triggered by a cascade of activation of enzymes called caspases. Caspases can be activated by extracellular or intracellular signals.
CHAPTER 7 FREESTANDING PRACTICE QUESTIONS
1. Kartegener’s syndrome is a rare genetic disorder that results in immotile cilia. The immotile cilia cause infertility, bronchiectasis (permanent dilation of the bronchi) and recurrent sinusitis (due to the inability to “push out” bacteria and particles from the sinuses). The genetic defect causes a deficiency of a protein primarily involved in which of the following structures?
A) Microfilament
B) Intermediate filament
C) Microtubule
D) Plasma membrane
2. Different proteins have been found to be involved in vesicular trafficking. Specific proteins are responsible for specific pathways; for instance, COP-I is responsible for retrograde transmission of vesicles, while COP-II is involved in anterograde transmission. Clathrin is involved in receptor-mediated endocytosis. Vesicles on which of the following pathways would be expected to have COP-I proteins on its surface?
A) RER → cis Golgi
B) RER → trans Golgi
C) cis Golgi → RER
D) Nucleus → RER
3. The steroid hormones produced by the smooth endoplasmic reticulum would be stored in which of the following organelles?
A) Secretory vesicles, so the hormones could be released when needed.
B) Peroxisomes, so the hormones could aid in lipid breakdown.
C) Lysosomes, so the hormones could aid in the destruction of excess secretory products.
D) Steroid hormones are not stored, as they are able to diffuse through lipid bilayers.
4. Which of the following gives the correct order for the signals leading to the formation of cyclic AMP? (GPCR = G-Protein Coupled Receptor)
A) Epinephrine → G-proteins → GPCR → adenylyl cyclase → cAMP
B) Epinephrine → GPCR → G-proteins → adenylyl cyclase → cAMP
C) Epinephrine → GPCR → adenylyl cyclase → G-proteins → cAMP
D) Epinephrine → adenylyl cyclase → cAMP → GPCR → G-proteins
5. The nuclear membrane is absent in which of the following phases of mitosis?
I. Anaphase
II. Telophase
III. Metaphase
A) I
B) I and II
C) I and III
D) II and III
6. Both the Golgi complex and rough endoplasmic reticulum contribute to protein modification through all of the following EXCEPT:
A) phosphorylation.
B) creation of disulfide bridges.
C) glycosylation.
D) creation of peptide bonds.
7. Tumor suppressor genes like BRCA1/2 promote anticancer processes like DNA repair. When these genes are disabled via mutation, the risk for developing cancer later in life increases dramatically. A female who inherits a single mutation in BRCA2 has a 50% and 20% chance of developing breast and ovarian cancer, respectively. Which of the following best explains why these respective chances are not 100%?
A) Individuals who are homozygous for the BRCA2 mutation do not have a functional allele.
B) Individuals who are homozygous for the BRCA2 mutation can still rely on other DNA repair mechanisms.
C) Individuals who are heterozygous for the BRCA2 mutation still have a functional allele.
D) Individuals who are heterozygous for the BRCA2 mutation can still rely on other DNA repair mechanisms.
CHAPTER 7 PRACTICE PASSAGE
Apoptosis is the most studied form of programmed cell death. It is limited to multicellular organisms and is a process that involves cells being dismantled from within. When a cell is undergoing apoptosis, it has several characteristic morphological traits; for example, the cells retract and become rounded as they detach from the extracellular matrix and neighboring cells. In addition, the plasma membrane starts blebbing, and apoptotic bodies (similar to vesicles) are pinched off the plasma membrane.
There are several stimuli that could induce apoptotic pathways in a cell. For example, there are receptor-mediated or extrinsic signals that trigger a cell to initiate apoptosis (Figure 1).
Figure 1 The Extrinsic Apoptosis Pathway
In this case, a death receptor ligand such as FasL or TNFα binds to a death receptor on the surface of the cell. This causes recruitment of adaptor proteins (such as FADD, or Fas-associated death domain protein) to the plasma membrane. The adaptor proteins serve as docking sites for other proteins, most commonly caspase 8, which is recruited to the plasma membrane, aggregated, auto-processed and ultimately activated. Active caspase 8 is a protease and cleaves other members of the caspase family (such as caspase 3, 6, or 7) and a protein called BID. The cleaved form of BID (tBID) promotes the release of cytochrome c from mitochondria, causing apoptosome assembly and downstream activation of other caspase family members. These proteins dismantle the cytoskeleton and cause nuclear fragmentation, detachment from the surrounding environment, genome degradation, and fragmentation of organelles such as the ER, Golgi, and mitochondria. After termination of the demolition phase, phagocytes are recruited to the apoptotic cell to clean up the cellular debris.
Autophagy is also a self-destructive process, but does not necessarily lead to cell death. When cells are under nutrient stress, reactive oxygen stress, or organelle stress, parts of the cytoplasm and intracellular organelles are sequestered in specialized double-membrane organelles called autophagosomes. These vacuoles ultimately fuse with lysosomes (to form autolysosomes) and the acid hydrolases in the lysosome degrade the contents, which are then recycled to the cell to provide the missing or limited nutrients.
In many situations, if the degradation of cellular material does not alleviate the stress that induced autophagy, the cell will switch to apoptosis. The signaling axis that controls the switch between these two pathways is complex and has been difficult to elucidate. It is now known that autophagy gene 5 (or Atg5) is an important and essential protein in autophagy induction and in the regulation of apoptosis. For example, Atg5 generally induces autophagy by promoting autophagosome and autolysosome formation. Atg5 also enhances the susceptibility of cancer cells to apoptosis. In cell culture experiments where Atg5 expression is decreased, autophagy is abolished and apoptosis incidence is reduced. It has been found that the full-length Atg5 protein induces autophagy. However, upon lethal stress, the full-length Atg5 is proteolytically cleaved to generate a 24 kDa pro-apoptotic protein. The truncated Atg5 translocates to the mitochondria and promotes cytochrome c release and mitochondrial permeabilization.
1. p53 is a tumor suppressor protein that is commonly mutated or deleted in aberrantly growing cancer cells. Which of the following is the most likely function of p53?
A) It is a transcription factor that induces expression of pro-apoptotic proteins.
B) It indirectly induces autophagy to supply the cell with biomacromolecules.
C) It stimulates phospholipid synthesis in the smooth ER.
D) It induces cell cycle progression in response to genome stability, as part of the G2-M checkpoint pathway.
2. Which of the following are NOT examples of enzymes found in lysosomes?
A) Phosphatases and nucleases
B) Phospholipase C and adenylyl cyclase
C) Proteases and peptidases
D) Glucosidases and lipases
3. Which of the following would NOT induce autophagy?
A) Increase in inner mitochondrial membrane permeability
B) Accumulation of incorrectly folded proteins in the ER lumen
C) Incomplete genome replication
D) Decreased intracellular metabolite concentrations
4. Cytochrome c :
I. is located in the inner mitochondrial membrane
II. undergoes redox reactions
III. functions in anaerobic respiration
A) I only
B) III only
C) I and II
D) I, II and III
5. A researcher adds the BAX channel inhibitor Bci1 to a plate of cells in the lab. Which of the following is observed?
A) Retention of cytochrome c in the inner mitochondrial membrane
B) Inhibition of BAX homodimer formation in the inner mitochondrial membrane
C) Rounded cells and blebbing of the plasma membrane
D) A rapid increase in the amount of cellular autophagy
6. Monophosphate Atg13 initiates autophagosome formation and mTOR phosphorylates three amino acids on Atg13 simultaneously. Which of the following is true?
A) mTOR is anti-autophagic and functions as a kinase.
B) mTOR activates Atg13 and functions as a phosphatase.
C) Rapamycin, an mTOR inhibitor, represses autophagy.
D) In cells that are undergoing autophagy, Atg13 is phosphorylated on three amino acids.
7. Which of the following is true of Atg5?
A) Southern blot analysis of proteins from healthy cells would show a 24 kDa Atg5.
B) Calpain, the protease that cleaves Atg5, is activated in cells that have bound FasL.
C) Western blot analysis of proteins from cells undergoing autophagy would show a 19 kDa Atg5.
D) Healthy cells and autophagic cells have alternative splicing of Atg5 to generate two different proteins.
SOLUTIONS TO CHAPTER 7 FREESTANDING PRACTICE QUESTIONS
1. C Cilia and flagella are both made up of microtubules; thus, a defect causing immotile cilia would likely involve a protein associated with microtubules (choice C is correct). In fact, Kartegener’s syndrome is a defect in the dynein protein, which is an ATPase that links the peripheral 9 doublets of the cilium (remember that eukaryotic cilia and flagella both have a 9 + 2 arrangement of microtubules) and causes bending (motion) of the cilium by differential sliding of the doublets. Microfilaments are composed of the protein actin and are responsible for contractile motility and changing of cell shape, specifically, muscle contraction, leukocyte motility, and cytokinesis (choice A is wrong). Intermediate filaments are primarily responsible for the cytoskeletal structure of cells (choice B is wrong). The plasma membrane is composed of phospholipids and cholesterol, although it does include many different proteins. However, the proteins of the plasma membrane are not responsible for cilia motility (choice D is wrong).
2. C The question stem states that COP-I proteins are involved in retrograde, or “backward,” vesicular trafficking. Therefore, these vesicles would be traveling in the reverse pathway. Choice C (cis Golgi to RER) is the only correct option listed. The anterograde, or “forward,” pathway in cells involves vesicles traveling from the RER to the cis Golgi (choice A is wrong), the medial Golgi, the trans Golgi and then to the cell membrane or other target site within the cell. Vesicles do not normally travel from the RER to the trans Golgi (choice B is wrong). Also vesicles are not involved in transport from the nucleus (choice D is wrong).
3. D Because steroid hormones are made from cholesterol as lipid derivatives, they cannot be stored by lipid bilayers. Since secretory vesicles, peroxisomes, and lysosomes are membrane-bound, they cannot store steroids (choices A, B, and C are wrong). Note also that peptide hormones are stored in secretory vesicles (choice A would be true of a peptide hormone, but not of a steroid hormone). Be careful to differentiate between answer choices that are simply true versus answer choices that actually address the question. Though choices A, B, and C all describe the general function of their particular organelle, only choice D addresses the issue specific to steroids.
4. B Since the question is asking about the formation of cAMP, it should be the last step in the sequence (choice D can be eliminated). G-protein coupled receptors receive the signal from ligand binding (in this example, epinephrine), and affect G-proteins. Therefore, GPCR must come before G-protein in the sequence (choice A can be eliminated). GPCRs activate G-proteins, which then affect the enzyme adenylyl cyclase (choice B is correct and C is wrong). Remember also that some GPCRs activate adenylyl cyclase and others inhibit it. This depends on the cell, the ligand, and many other factors.
5. C Item I is true: the nuclear membrane is degraded by the end of prophase so that the chromosomes can be appropriately separated into the daughter cells. It is therefore absent in anaphase (choice D can be eliminated). Item II is false: The nuclear membrane reforms after cytokinesis in telophase (choice B can be eliminated). Item III is true: Since the membrane does not reform until telophase, it must be absent in metaphase (choice A can be eliminated).
6. D Phosphorylation, formation of disulfide bridges, and glycosylation are all types of protein modification and can occur in both the Golgi apparatus and the rough ER (choices A, B, and C can be eliminated). Peptide bond formation occurs during translation, prior to any protein modification (choice D is not a type of protein modification and is the correct answer choice).
7. C A female who inherits a single mutation still has an allele which can produce functional proteins (choice C is correct) and is heterozygous, not homozygous (choices A and B are wrong). While an individual who is heterozygous for the mutation can rely on other DNA repair mechanisms, this is not the primary reason why these individuals will not have a 100% chance of developing breast or ovarian cancer (choice C is better than choice D).
SOLUTIONS TO CHAPTER 7 PRACTICE PASSAGE
1. A The key to answering this question is decoding the question stem: If p53 is commonly defective in cancer cells, it must normally have some sort of protective role. In other words, p53 loss is beneficial for cancer cells. If p53 normally induces apoptosis and becomes mutant or lost, then it would no longer cause cell death in aberrant conditions. This could be a benefit for cancer cells, and in fact could lead to the rapid cell proliferation and tumor growth that is typically seen in cancer (choice A is correct). The passage says that autophagy can help the cell deal with times of stress, including nutrient deprivation. Since cancer cells grow quickly, they have high metabolic demands, which are accommodated in part using autophagy to supply macromolecules. If p53 normally helped the cell supply macromolecules, p53 loss would not help the growth of cancer cells (choice B can be eliminated). Similarly, if cancer cells are growing quickly, they will need lots of phospholipids. If p53 normally stimulated phospholipid synthesis, its loss would not be beneficial for the cancer cell (choice C can be eliminated). Finally, if p53 normally pushed the cell cycle forward, its loss would arrest the cell cycle. This is clearly not the case in “aberrantly growing cancer cells” (choice D is wrong).
2. B The passage says that the lysosome contains acid hydrolases, which degrade the contents of the autophagosomes. Since the autophagosomes contain cellular and organelle material, it is possible that they would contain proteins (which would be broken down by proteases and peptidases; choice C can be eliminated), nucleic acids (which would be broken down by nucleases; choice A can be eliminated), carbohydrates (which would be broken down by glucosidases) and lipids (which would be broken down by lipases; choice D can be eliminated). However, phospholipase C and adenylyl cyclase are both involved in cell signaling pathways, downstream of ligands binding receptors. Neither of these would function in macromolecule degradation (choice B would not be found in lysosomes and is the correct answer choice). Note that phosphatases remove the phosphate group from molecules, and in many cases, this is part of degradation.
3. C The passage says that autophagy is induced when cells are under nutrient stress, reactive oxygen stress, or organelle stress. Decreased intracellular metabolite concentrations would be an example of nutrient stress (choice D would induce autophagy and can be eliminated). An increase in mitochondrial permeability and an accumulation of incorrectly folded proteins in the ER lumen are examples of organelle stress (autophagy degrades broken or sickly organelles; choices A and B could induce autophagy and can be eliminated). While incomplete genome replication in the S phase is a problem for cells, there is no information to indicate that this would induce autophagy. Also, the effects of autophagy as described in the passage (break down of cytoplasmic and organelle material) would not solve this problem since the genome cannot be degraded (there is only one, after all). In fact, incomplete genome replication is detected by cell cycle checkpoint pathways to halt cell cycle progression and allow time for DNA replication and repair (choice C would not induce autophagy and is the correct answer choice).
4. C Item I is true: Figure 1 shows that cytochrome c is normally located in the inner mitochondrial membrane (choice B can be eliminated). Item II is true: Cytochrome c is part of the electron transport chain and therefore undergoes redox reactions (choice A can be eliminated). Item III is false: The electron transport chain is a major component of aerobic respiration, not anaerobic respiration (choice D can be eliminated and choice C is correct).
5. A According to the figure, BAX proteins form channels in the mitochondrial membrane and this facilitates cytochrome c release. If Bci1 inhibits these BAX channels, it would cause cytochrome c to remain in the inner mitochondrial membrane (choice A is correct). This would prevent apoptosis (and thus rounded cells and blebbing; choice C is wrong). While choice B might be tempting, just because Bci1 inhibits BAX channels doesn’t mean it has to inhibit homodimer formation (although this is certainly possible). We can be sure about the end result, though (retention of cytochrome c), even if we aren’t sure of the mechanism (choice A is better than choice B). Note that BAX functions in apoptosis, not autophagy (choice D is wrong).
6. A Note that there is no information about Atg13 in the passage, so this question must be answered based entirely on information in the question. The question text states that when Atg13 is phosphorylated on one amino acid, it initiates autophagy by inducing autophagosome formation (choice D is wrong). If mTOR phosphorylates Atg13 on three amino acids, it is inhibiting Atg13 function and is inhibiting autophagy (choice A is correct and choice B is wrong). If rapamycin inhibits mTOR, it is inhibiting an inhibitor of autophagy. In other words, rapamycin would induce autophagy (choice C is wrong). A kinase phosphorylates molecules (using ATP as the source of the phosphate) and a phosphatase removes phosphate groups, although this information is not needed to answer the question.
7. B The passage says that Atg5 cleavage (from a longer precursor protein to a shorter 24 kDa protein) is involved in the switch from autophagy to apoptosis. If cells have bound FasL (a death receptor ligand), they are undergoing apoptosis, or will be soon. Since Atg5 cleavage induces apoptosis, choice B is very likely. Southern blot analysis is a lab technique used to study DNA, not proteins (choice A is wrong). While western blot analysis is the correct lab technique to study proteins, in cells undergoing autophagy (not apoptosis), Atg5 would be in the longer, nontruncated form (the 24 kDa protein, not the 19 kDa protein; choice C is wrong). The passage describes proteolytic cleavage as the mechanism for generating differently sized Atg5, not alternative splicing (choice D is wrong).
1 It could be either. mRNA and translation are found in the cytoplasm of both prokaryotes and eukaryotes, so the cell could be either.
2 The isolated genome and the genome in the cell respond differently, so the key is not the circular or linear nature of the genome (B and D are wrong). The key is that in prokaryotes the injected cytoplasmic DNase will have access to the genome to degrade since they are in the same compartment, while in eukaryotes the DNase will not have access to the genome unless it enters the nucleus (C is the best choice).
3 DNA polymerase cannot synthesize DNA without an RNA primer and cannot synthesize DNA in a 3 to 5 direction. It will replicate DNA from an RNA primer at (or very near to) the end of the chromosome, but the RNA primer cannot be replaced with DNA. With each round of replication, the chromosome grows shorter and shorter because of the inability to synthesize DNA at its very ends. Because of the large number of repeated sequences at the ends (the telomeres) the loss of a little bit of DNA is usually not critical. However, eventually all the telomere sequences are lost, and gene sequences start to be lost. If the lost gene sequence is critical for cell function, the cell will die at this point. Telomerase helps prevent this problem. This unique enzyme contains an RNA sequence and acts as a reverse transcriptase, utilizing this RNA sequence as a template to extend the DNA at the end of the chromosome. This provides a location where a normal primer can be synthesized and DNA replication can proceed along the very end of the chromosome, preserving it. Note that telomerase is turned off in most cells, and its inactivity is implicated in cell aging and death.
4 The retroviral genes will not be expressed very frequently, and the virus will tend to remain as a provirus unless a change in the surrounding heterochromatin allows viral genes to be expressed.
5 The nucleolus is largest in cells that are producing large amounts of protein. The increased size reflects increased synthesis of ribosomes.
6 The DNA will serve as template for ribosomal RNA production.
7 No. Bacteria have only a single kind of RNA pol which is responsible for all transcription.
8 No, it is not. The space between the nuclear membranes is contiguous with the ER lumen, which is isolated from the cytoplasm.
9 Yes. The protein is small enough that it can still pass through the nuclear pores by diffusion even without a nuclear localization sequence.
10 Aminoacyl tRNA synthetases are enzymes that function in the cytoplasm to attach amino acids to their respective tRNAs. They are never needed in the nucleus and would not be found there (choice D is correct). The protein components of ribosomes are synthesized in the cytoplasm and then imported into the nucleus to be assembled in the nucleolus (choice A would be found in the nucleus and can be eliminated). Splicing occurs in the nucleus, so anything involved in splicing would be found there (choice B can be eliminated). Histones are used for DNA packaging and would be found in the nucleus (choice C can be eliminated).
11 Gold beads are not normally found in cells, so there cannot be an existing mechanism for moving them. However, since the cell is capable of moving them when the localization signal is attached, the localization signal must be somewhat non-specific (choice C is correct). It is true that the nuclear localization signal is lysine-rich, but this cannot be concluded based on the given information (true, but doesn’t answer the question, choice A is wrong). If gold beads had an inherent import signal, then they would be transported into the nucleus on their own, without the researcher having to bind them to the localization sequence (choice B is wrong). If simple diffusion were the primary means of moving things into the nucleus, no import signal would be needed (choice D is wrong).
12 The folding of the membrane increases its surface area and allows for increased electron transport and ATP synthesis per mitochondrion. (Folding is used elsewhere to increase surface area, such as in the kidney tubules and the lining of the small intestine.)
13 No, in the cytoplasm.
14 Pyruvate is transported through the inner mitochondrial membrane by a specific protein in the membrane.
15 Remember that bacterial electron transport depends on a proton gradient across the cell membrane. In a Gram-negative bacterium, this membrane would correspond to the mitochondrial inner membrane.
16 The coding system of the cellular genome is different from that of the mitochondrial genome. One might wonder how our transcription and translation machinery could sensibly produce mitochondrial gene products.
17 All of her children will have it, since they will inherit mitochondria exclusively from her. For a maternally inherited trait, it doesn’t matter whether the father has it or not.
18 The only way a protein can be smaller than would be expected from its mRNA would be if some post-translational modification were to occur (choice D is correct). Choices B and C are pre-translational modifications and would not account for a size difference between mRNA and protein, and since secreted proteins are synthesized on the rough ER, they are inaccessible to cytoplasmic proteases (so A is wrong).
19 The cellular exterior
20 Yes. Remember, the ER lumen is equivalent to (contiguous with) the extracellular space.
21 No. It is the signal peptide in the nascent polypeptide that is recognized and bound by SRP and taken to receptors on the surface of the rough ER. The signal is an amino acid sequence on the nascent polypeptide, not a nucleotide sequence on mRNA.
22 Note that cis means “near,” as in a cis double bond. Trans means “far.” Medial means “in the middle.” Also note that the order is alphabetical: cis-medial-trans.
23 No. Secretory proteins must proceed via a specific path: from the ER to the cis Golgi to the medial and trans Golgi and from there to the cell surface.
24 Lipid bilayers form spontaneously, as the lowest energy state, without external energy input. This describes a process with a negative ∆G.
25 C. Phospholipids have hydrophobic components (fatty acid acyl chains) and hydrophilic components (phosphate and choline, for example, in phosphatidyl choline).
26 It occurs in the rough ER as the protein is translated and threaded across the ER membrane.
27 After a short period of time, the red and green tagged lipids will diffuse laterally and mix. An even distribution of the tags will be seen across the surface of the new hybrid cell.
28 Unsaturated fatty acids, with a kinked structure, have fewer van der Waals interactions, and therefore allow a more fluid membrane structure. Increasing the unsaturated fatty acids will increase membrane fluidity.
29 The addition of a solute to a liquid always results in the effects of all the colligative properties simultaneously. Therefore, choice D is the answer.
30 Remember, ∆G = ∆H – T∆S, so increasing ∆S decreases ∆G, indicating a thermodynamically favorable process.
31 Water will flow into the cell through the plasma membrane until the cell volume increases to the point that the cell bursts.
32 hydrophobicity
33 No. Ion channels are only involved in facilitated diffusion, the movement of molecules down an elecrochemical gradient with the help of a protein.
34 No. Porins are large holes, and ion channels are small, usually regulated channels. If porins and ion channels were found in the same membrane, the ion channels would be useless, because ions would flow in an unregulated manner through the pores.
35 Facilitated diffusion is the movement of molecules down a gradient with the help of a protein (choice B is correct). Membrane proteins are not required for simple diffusion (choice A is wrong), and active transport involves moving things against their gradients (choices C and D are wrong). Note also that in secondary active transport, the ion movement down its gradient must be coupled to the movement of some other molecule against its gradient.
36 The pumping of ions against a gradient which is coupled to ATP hydrolysis is primary active transport.
37 The Na+/K+ ATPase is required to establish the resting membrane potential in which the cellular interior has a negative charge. It pumps out one net positive ion. If this net positive ion stays inside the cell, the resting potential becomes less negative (choice B is correct). Since the interior of the cell is now more charged, the cell will have a tendency to take on water by osmosis, and will swell (choice A is wrong). Secondary active transport depends on the gradient established by primary active transport (the Na+/K+ pump). If the pump is shut down, the gradient won’t be established, and secondary active transport will also stop (choice C is wrong). The Na+/K+ ATPase has nothing to do with cellular proliferation (choice D is wrong).
38 The pump would run backwards! Remember: All active transporters are reversible.
39 The cell contains millions of negative charges on macromolecules (e.g., nucleic acids). For the charge to be “approximately” balanced on both sides of the membrane, some negatively-charged substance must be more concentrated outside. Chloride serves this role. Why did we say “approximately” balanced? Remember: The cell is a bit more negative on the inside; that’s the RMP.
40 No. Clathrin is a fibrous protein inside the cell that associates with the cytoplasmic portions of the cell-surface receptors that bind lipoproteins.
41 Both import a particular substance. One difference is that in endocytosis the substance ends up sealed in an endosome, whereas in active transport the substance is just dumped into the cytoplasm.
42 sperm
43 An epithelial cell layer is a layer of cells which lies “upon nipples” of a type of extracellular connective tissue called basement membrane (epi- means “upon,” and -thele means “nipple,” in the sense of small bump). The basement membrane is a strong molecular sheet made of collagen. Under the microscope the basement membrane under epithelial cells has “bumps” which make epithelial cell layers easy to recognize.
44 No. It is free to move around on the apical surface, but the tight junctions prevent it from diffusing to the basolateral surface.
45 A mnemonic is “I Pee on the MAT,” where I is for interphase.
46 Presumably so that they can be separated without tangling.
47 This stage of prophase is also referred to as “prometaphase.” It is the last event in prophase and is rather dramatic; once the nuclear membrane is disintegrated into vesicles, the spindle fibers can attach to the centromeres of the chromosomes and the cell can enter metaphase.
48 a ring of microfilaments encircling the cell and contracting
49 If a chromosome had more than one centromere, it could be pulled toward different ends of the cell simultaneously and be torn (choice D is correct). Each eukaryotic gene has only one reading frame, but since there are many genes per chromosome there are different reading frames, too. Note that the total number of possible reading frames is only 3, since a codon is only 3 nucleotides long (choice A is wrong). Eukaryotic chromosomes are so large that they must have more than one origin of replication to finish replication of the genome in a reasonable time period (choice B is wrong), and each gene has its own promoter, and there are many genes per chromosome (choice C is wrong).
50 If the genome is not completely replicated and condensed prior to mitosis, it will be torn during cell division. Each daughter cell will receive only pieces of the genome rather than the complete genome and will not be able to survive (choice B is correct and choice A and C are wrong). DNA repair systems can only repair sequence errors or minor structural problems; this problem would be too large to fix (choice D is wrong).
51 Teratomas form when oncogenes cause certain tissues to dedifferentiate (lose their specific function and regress), and then redifferentiate to become something different. This is why teratomas can contain tissues such as teeth and hair.
52 Since regenerative capacity involves cell growth, dedifferentiation and redifferentiation, the possibility exists for the process to go awry and for cancerous growth to then be stimulated. Regeneration needs to be tightly regulated in order for it to proceed correctly.