CHEMICAL BIOLOGY
Pre-mRNA splicing
John Karijolich and Yi-Tao Yu, University of Rochester Medical Center, Rochester, New York
doi: 10.1002/9780470048672.wecb520
Pre-mRNA splicing is an RNA processing reaction by which introns are removed from an mRNA precursor and exons are precisely joined together to form a functional mature mRNA. Splicing involves two successive trans-esterification reactions that occur in the spliceosome, which is a multicomponent complex composed of a large number of protein factors and five small nuclear RNAs (snRNAs), each functioning as an RNA-protein complex (ribonucleoprotein or snRNP). A significant body of evidence indicates that both spliceosome assembly and catalytic core formation are orchestrated by an intricate network of RNA-RNA interactions. Most eukaryotic introns invariably start with the dinucleotide GT (GU in RNA) and end with the dinucleotide AG. Splicing of these introns occurs in the major spliceosome containing five abundant snRNPs (U1, U2, U4, U5, and U6). In high eukaryotes, a rare class of introns also exists, beginning with the dinucleotide AT (AU in RNA) and ending with the dinucleotide AC. Removal of this class of introns requires the participation of a different set of snRNPs, namely U11, U12, U5, U4atac, and U6atac. Recent advances in pre-mRNA splicing have provided strong evidence for an RNA enzyme catalyzing the two trans-esterification reactions in the spliceosome.
Introduction
It was discovered in 1977 that several adenovirus genes (in the form of DNA) contained intervening sequences that were not present in their mature mRNAs (1, 2). Shortly thereafter, other eukaryotic genes were similarly found to contain intervening sequences that disrupt the real hereditary message. Thus, the intervening sequences were named introns and the protein coding sequences (including the 5' and 3' untranslated) were termed exons (3). Since the discovery of introns, the phenomenon of intron-containing genes (often referred to as “split genes”) has been found to be widespread in eukaryotes. In fact, almost all genes in high eukaryotes contain introns (the exceptions are those encoding histones and interferons). Even in budding yeast, more than 250 genes contain introns. Early work on RNA metabolism in eukaryotes identified a large number of short-lived heterogeneous nuclear RNAs (hnRNAs) that are much longer than mature mRNAs in the cytoplasm. The hnRNA sequence can form a perfect double-stranded RNA-DNA duplex when hybridized with its own gene (4). Taken together, these observations produced the realization that transcription of a gene initially produces a perfect copy of RNA known as mRNA precursor (pre-mRNA or hnRNA) containing not only exons but also introns. Introns must be removed from the precursor before the resulting mature mRNA is transported to the cytoplasm where it directs the translation of protein. The removal of introns from mRNA precursor is catalyzed by an RNA processing reaction known as pre-mRNA splicing. Pre-mRNA splicing occurs in the spliceosome, a massive protein-RNA complex consisting of a large number of proteins and five small nuclear RNAs (U1, U2, U4, U5, and U6 snRNAs), each of which functions as a small nuclear ribonucleoprotein (snRNP) (5-8). Over the years, the spliceosomal snRNAs (or snRNPs) have been extensively studied and their roles in pre-mRNA splicing have been well established. A fairly detailed picture has emerged in which intricate networks of interactions among the components of the spliceosome, in particular the RNA-RNA interactions, orchestrate the assembly of the spliceosome as well as the formation of the catalytic center for catalysis (chemical reaction of splicing) (6, 9-11). Here, we discuss spliceosome assembly and pre-mRNA splicing, with an emphasis on the U snRNAs as well as some important spliceosomal proteins (both snRNP proteins and non-snRNP proteins).
Pre-mRNAs Contain Conserved Sequence Elements at the Splice Sites
In the late 1970s and early 1980s, the sequences of many eukaryotic genes became available for study, thus making it possible to conduct a comparative analysis to identify important sequence elements in pre-mRNA. Careful inspection of higher eukaryotic genes revealed several consensus sequences in introns at or near the 5' and 3' splice sites (12, 13). The 5' splice site consensus sequence is G/GURAGU (/ represents the 5' exon-intron junction; R depicts a purine; the underlined dinucleotide GU is invariant). The 3' splice site is YAG/G (here, / represents the 3' intron-exon junction; Y is a pyrimidine; the underlined dinucleotide AG is invariant), which is frequently preceded by a CU-rich region. In addition to the 5' and 3' splice sites, there is another consensus sequence, namely the branch site YNYURAC (N can be any nucleotide and the underlined adenosine is invariant) located 20-40 nucleotides upstream of the 3' splice site (Fig. 1). In budding yeast, the three consensus sequences at the 5’ and 3’ splice sites (including the branch site) have also been identified; however, there is no or a less-conserved CU-rich region preceding the 3' splice site. In contrast to the loosely defined consensus sequences in high eukaryotic pre-mRNAs, the sequence elements in yeast pre-mRNA are almost absolutely conserved: G/GUAUGU at the 5' splice site, CAG/G at the 3’ splice site, and UACUAAC at the branch site. Mutation of any invariant nucleotides within the consensus or conserved elements results in a total inhibition or a decreased level of pre-mRNA splicing in both yeast and higher eukaryotes, further confirming their importance in the splicing process. The question that then develops is what role do these consensus sequences (and the invariant nucleotides) play during splicing? Are they recognized by other trans-acting cellular components, and if so, what are they?
Early Recognition of the 5' Splice Site and the Branch Site
Although identified and characterized in the 1960s (14, 15), U1 and U2 snRNAs had not been assigned functions until the discovery of split genes (or pre-mRNA splicing). In an attempt to link U1 to pre-mRNA splicing, researchers first analyzed the sequence complementarity between the snRNA and the conserved sequence elements in the pre-mRNA. Excitingly, the sequence analyses revealed a striking complementarity between the evolutionarily invariant 5' end of U1 and the pre-mRNA consensus sequence at the 5' and 3' splice sites (16, 17). Although the original proposals for the involvement of U1 snRNA in splicing suggested that the 5' end of U1 may base-pair with consensus sequences surrounding the 5' and 3' splice site, in 1981 Mount and Steitz modified the model after observations that certain sequences in U1 predicted in binding the 3’ splice site were not conserved (18). Driven by this hypothesis, researchers carried out a series of experiments to confirm the involvement of U1 in pre-mRNA splicing. For instance, in 1983 the Steitz lab provided the first evidence that U1 snRNA’s primary role was the recognition of the 5' splice site (19). Using an in vitro transcribed RNA fragment of the major mouse P-globin gene, which contains an intron, they localized the U1 snRNP binding site by T1 ribonuclease treatment of the snRNP-RNA complex, immunoprecipitation of the resulting trimmed complex with anti-U1 antibodies, and T1 ribonuclease fingerprinting of the immunoprecipitate. Fingerprinting analyses revealed three oligonucleotides present: CAG, UUG, and UAUCCAG. All three of these fragments can be found together in a single region that encompasses the 5' splice site (GGGCAG/GUUGGUAUCCAG) (19). In a separate experiment, the 5’ end region of U1 was destroyed by oligodeoxynucleotide-directed RNase H digestion, resulting in a loss of splicing activity (20). In addition, an elegant genetic suppression assay was used to verify the base-pairing interaction between U1 and pre-mRNA (21). In that work, a point mutation was first introduced into the 5’ splice site region of a pre-mRNA. As expected, splicing of the pre-mRNA was inhibited. However, when a compensatory point mutation was introduced into U1 snRNA at the site (within the 5’ end region) that was predicted to restore the base-pairing interaction, splicing activity was indeed rescued (21). More recently, psoralen cross-linking also detected base-pairing interaction between U1 and pre-mRNA (22). Along the same line, U1 was physically detected in the splicing complexes (see below). Such an enormous amount of data has clearly demonstrated that, indeed, U1 recognizes pre-mRNA during splicing and that the recognition involves a direct base-pairing interaction between the U1 5’ end region and the pre-mRNA 5' splice site (see Fig. 1). Binding of U1 as well as other splicing factors to the 5' splice site of a pre-mRNA leads to the formation of an early complex, termed a commitment complex, that commits pre-mRNA to the splicing pathway (see below, Spliceosome Assembly Pathway) (23-28).
Similar experimental approaches were used to demonstrate the involvement of U2 snRNP in pre-mRNA splicing as well (29). A large body of evidence accumulated over the years unequivocally shows that U2 snRNP participates in pre-mRNA splicing. More specifically, U2 recognizes the pre-mRNA branch site via a specific base-pairing interaction involving a highly conserved region of U2 and the branch site consensus sequence (22, 30-35) (Fig. 1). This interaction specifically bulges out the branch point nucleotide adenosine, a configuration favorable for the in-line nucleophilic attack at the 5’ splice site (the first step of splicing) (see below).
Besides U1 and U2, many splicing factors (including snRNP proteins), such as SF1, SF3 (including p14, a subunit of SF3b), and p220 (Prp8p in yeast), among others, have also been implicated in participating or facilitating 5’ splice site and/or branch site recognition (36). For example, although the 5' splice site is initially recognized by the U1 snRNP via a base-pairing interaction, this interaction is stabilized by the U1-70K and U1-C proteins (U1 snRNP proteins), as well as members of the SR protein family (37) that bind to the 5' splice site before the entry of U1 snRNP. SR proteins contain a domain rich in arginine-serine (RS) dipeptides, and have numerous roles in pre-mRNA splicing, spliceosome assembly, and splice site recognition (38). In the case of the U2 snRNP interaction with the branch site, it is facilitated by splicing factors SF3a and SF3b (39), while its interaction is stabilized by Prp5, a spliceosomal ATPase (see below) (40).
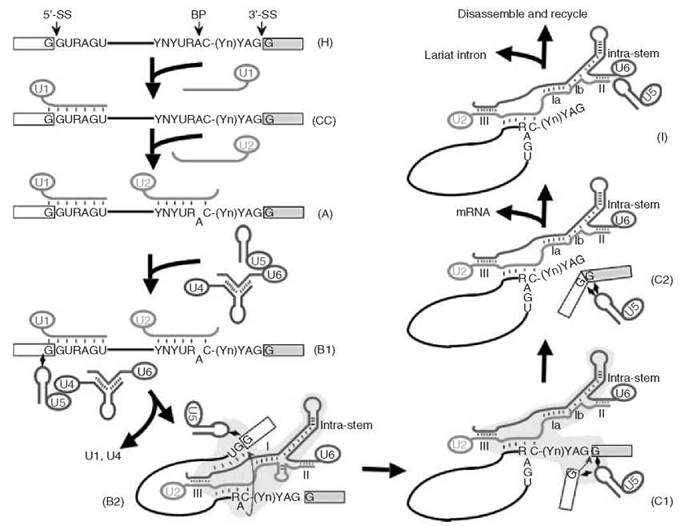
Figure 1. Major spliceosome assembly and action. First, U1 (red) assembles onto the pre-mRNA (in the H complex) to form a commitment complex (CC). Then U2 (green) joins, converting complex CC to complex A. Subsequent joining of the U4-U6-U5 (blue-brown-purple, respectively) tri-snRNP forms the major spliceosome (complex B1). The newly assembled spliceosome then undergoes a dynamic rearrangement of RNA-RNA interactions resulting in the release of U1 and U4 and the formation of complex B2. Formation of complex B2 triggers the first catalytic step, the formation of the cut-off 5' exon and the lariat structure, thus leading to the formation of a new complex (complex C1). After additional conformational changes, step 2—the joining of 5' and 3' exons—occurs and complex C2 forms. mRNA is then released, resulting in the formation of complex I. Finally, the lariat intron is released and the spliceosome disassembles. The 5' splice site (5'-SS), the 3' splice site (3'-SS) and the branch point adenosine (BP) are indicated. The consensus sequence elements at the 5' and 3' splice sites and the branch site are shown (R, purine; Y, pyrimidine; Yn, the polypyrimidine tract; N, any nucleotide). The thick lines represent snRNA strands (in colors) or the intron (black), and the boxes are exons. The short thin lines between RNA strands represent Watson-Crick base-pairing interactions. The lightning bulbs depict non-Watson-Crick base-pairing interactions. The bulged-out branch point adenosine is pictured after the joining of U2. The blue arrows indicate the chemical reactions, trans-esterifications, or nucleophilic substitution reactions. The yellow areas in complexes B2 and C1 represent the catalytic center for catalysis. U2-U6 duplexes [Helices I (or la and Ib), II and III] in the spliceosome are shown. The U6 intramolecular stem structure (Intra-stem) is also pictured. The extended U6 intramolecular stem (before the first step of splicing, complex B2) or the U6 intramolecular stem plus U2-U6 Helix Ib (before the second step of splicing, complex C1), constitutes a structure that resembles domain V (active site) of group II introns (self-splice introns). Note: Different nomenclatures are used to describe splicing complexes in mammals and yeast. Here, the nomenclatures are adopted from the mammalian system (66).
Splicing Factors Recognize the 3' Splice Site
U2AF (U2-auxiliary factor) is one of several splicing factors that have been implicated in 3’ splice site recognition (36, 39). U2AF has two subunits, namely U2AF65 ( 65 KD) and U2AF35 (35 KD). It has long been known that U2AF65 binds to the polypyrimidine track upstream of the 3’ splice site of pre-mRNA in high eukaryotes (36, 39, 41). However, the role of U2AF35 had remained elusive until 1999 when a connection between U2AF35 and the dinucleotide AG at the pre-mRNA 3’ splice site was finally established (42-44). Using a variety of experimental approaches, including U2AF depletion and reconstitution, cross-linking and immunoprecipitation, Selex, nuclease protection, and mutational analysis, three research groups independently demonstrated that U2AF35 is actively recognizing the dinucleotide AG at the 3’ splice site during splicing (42-44).
Other splicing factors may also play important roles in 3' splice site selection. For example, using the yeast genetic approach and the mammalian in vitro splicing system, researchers demonstrated that the splicing factor Slu7 is important for the selection of the canonical AG at the 3' splice site during splicing (45, 46).
Splicing Occurs Via a Two-Step Trans-esterification Reaction Pathway
In the early 1980s, a mammalian in vitro splicing system (Hela nuclear extracts) was developed, leading to the discovery of two splicing intermediates, the cut-off 5' exon intermediate and the 2/3 lariat intermediate (intron-3' exon in a lariat form) (47, 48). Importantly, the lariat (or branched) structure was also identified in cellularly derived RNAs (49, 50). Soon after, the yeast in vitro system also became available, and it, too, allowed the detection of the cut-off 5’ exon and the 2/3 lariat intermediates (51). A two-step trans-esterification reaction pathway was thus established (Fig. 2). In the first step, the 2' hydroxyl group (2'-OH) of the branch point nucleotide adenosine attacks the phosphate at the 5' exon-intron junction (5' splice site), resulting in the cleavage of the phosphodiester bond between the 5’ exon and intron and the concurrent formation of a new 5’-2’ phosphodiester bond between the 5' end of the intron and the branch point adenosine. Thus, a lariat-structured intermediate (the 2/3 lariat intermediate) and the cut-off 5' exon intermediate are produced. In the second step, the 3'-OH group of the cut-off 5' exon attacks the phosphate at the intron-3' exon junction (3’ splice site), releasing the lariat intron product and generating the spliced mature mRNA product. According to this pathway, the branch point adenosine and the guanosines at the 5’ and 3’ ends of the intron are the key nucleotides as they directly participate in the chemical reactions. In fact, these three nucleotides are among the most conserved nucleotides in pre-mRNA (see above).
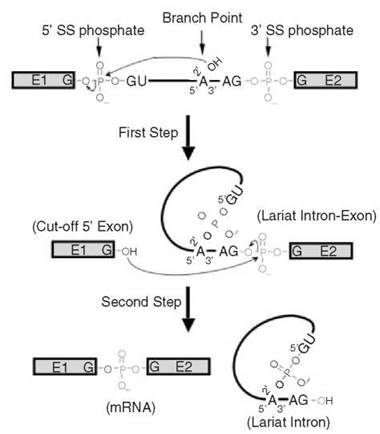
Figure 2. Details of two successive trans-esterification reactions. In the first step, the 2'-OH group of the branch point adenosine nucleophilically attacks the phosphate at the junction of the 5' exon and intron (5' splice site), resulting in the formation of a new 5'-2' phosphodiester bond between the first nucleotide of the intron and the branch point adenosine (lariat structure formation) and breakage of an old 3'-5' phosphodiester bond between the last nucleotide of the 5' exon and the first nucleotide of the intron (cut-off 5' exon formation). In the second step, the 3'-OH group of the cut-off 5' exon nucleophilically attacks the phosphate at the junction of the intron and 3' exon, ligating the two exons (mRNA formation) and releasing the lariat intron. The phosphates at the 5' splice site (red) and at the 3' splice site (green) and the branch point adenosine and its 2’-OH group are pictured. The lines represent the intron and boxes depict exons (E1 and E2).
It was anticipated that splicing occurs in a large RNA-protein complex (or the spliceosome), given that multiple snRNAs and proteins had been identified as functional components that participate in splicing. However, how big the spliceosome is and, perhaps more importantly, how it is assembled had remained largely unclear. The availability of the in vitro splicing systems offered a great opportunity to analyze splicing complexes. In 1985, using velocity gradient sedimentation, several research groups identified large complexes in which pre-mRNA, splicing intermediates and products, as well as U snRNAs were found (52-57). These complexes have a sedimentation constant of 50-60S when mammalian systems were used (53-57) or ~40S when the yeast system was used (52). Soon after, a more convenient and effective technique involving native gel electrophoresis was developed, leading to the identification of a number of splicing complexes assembled at different times during splicing in vitro (35, 58-62). Further, using biotinylated pre-mRNA, researchers were able to isolate splicing complexes through biotin-streptavidin affinity chromatography, allowing a systematic identification of the spliceosomal components in various splicing complexes (28, 63-65). A stepwise spliceosome assembly pathway was thus established (Fig. 1) (66, 67).
The assembly starts with a nonspecific H complex that forms as soon as the pre-mRNA is mixed with the splicing extracts (62, 66, 68, 69). This complex is then turned into a commitment complex (CC complex or the E complex) after binding with U1 snRNP and various protein factors, including SR proteins, U2AF (Mud2p in yeast), and SF1 (BBP/Msl5p in yeast) (23-28). The commitment complex is a complex that is committed to splicing out the intron on which it assembles. U2 snRNP then joins the commitment complex, converting it into a presplicing complex, namely complex A (27, 35, 58, 60-62, 66, 67, 69). After binding with the preformed functional U4/U6/U5 tri-snRNP (70-74), in which U4 and U6 are extensively base-paired with each other, complex A is then converted to complex B1 (61, 66, 67). Complex B1 undergoes a conformational change, and it becomes complex B2, which is now competent for the first step of splicing (61, 66, 67). After the first step of splicing and additional conformational changes, complex B2 is turned into complex C1 in which the second step of splicing occurs (61, 66, 67). Complex C1 then changes to complex C2, which contains spliced products (61, 66, 67). The mature mRNA is then released, changing complex C2 to complex I (62, 66, 67). Finally, the lariat intron product is released and complex I dissociates into its components, which subsequently participate in a new cycle of spliceosome assembly (66, 67).
Dynamic Rearrangements of RNA-RNA Interactions within the Spliceosome
Although kinetic analysis of spliceosome formation led to the discovery of a stepwise assembly pathway, the detailed network of interactions among the components within the massive spliceosome had remained unclear until the early 1990s when a series of cross-linking and yeast genetic suppression experiments were carried out to tackle this problem. Several cross-linking techniques, including psoralen cross-linking to detect Watson-Crick base-pairing interactions, 4-thioU crosslinking to detect non-Watson-Crick contacts between two nucleotides, and direct short wavelength UV cross-linking to detect various interactions, were used effectively to capture the interactions between U snRNA and pre-mRNA and among U snRNAs themselves (22, 75-79). Cross-linking was performed at various time points in parallel with splicing and spliceosome assembly assays, such that any detected cross-linking species could be assigned to a particular stage (splicing complex) during spliceosome assembly and splicing. In the meantime, sensitive yeast and mammalian genetic suppression assays also identified several novel RNA-RNA interactions (80-88). Recently, some of the RNA-RNA interactions within the spliceosome were further refined through the use of NMR spectroscopy (89). The large amount of data accumulated over the years provided a detailed picture revealing a network of RNA-RNA interactions within the spliceosome.
In addition to the early base-pairing interactions between U1 and the 5' splice site and between U2 and the branch site, a number of new RNA-RNA interactions were detected at late times during spliesome assembly, especially after the spliceosome is fully assembled (complex B1) (Fig. 1). Interestingly, the formation of some new interactions is accompanied by the dissociation of old interactions, indicating that dynamic rearrangements occur within the spliceosome. Specifically, entry of the U4/U6-U5 tri-snRNP to form complex B1 may be mediated at least in part through the base-pairing interaction between the 5' end of U2 and the 3' end of U6 (U2-U6 Helix II) (78, 82, 86). As soon as the U4/U6-U5 tri-snRNP enters the splicing complex, U5 snRNA interacts, in a non-Watson-Crick pairing manner, with the exon sequence at the 5’ splice site, and possibly with the exon sequence at the 3’ splice site as well (75-77, 83, 84, 87). In the meantime, a conserved sequence of U6 (ACA-GAGA) displaces U1 in interacting with the 5' splice site (79, 88). Concurrently, or immediately after, U6 dissociates from U4. The freed U6 sequence then forms new duplexes with U2 (Helices I and III, see Fig. 1) (22, 81, 85, 89), which are believed to be at least part of the catalytic center (9, 10). This dynamic rearrangement results in the release of both U1 and U4 from the spliceosome and the formation of an active catalytic center for the first step of splicing (complex B2). After the first chemical reaction, the spliceosome undergoes additional conformational changes, resulting in the disruption of the base-pairing interaction between U6 and the 5' splice site, the formation of U2-U6 Helix Ib, and the repositioning of the reaction substrates within the catalytic center (complex C1) (80, 89, 90). These conformational changes directly trigger the second step of splicing that generates the mature mRNA and lariat intron products (complex C2). After the second step of splicing, further conformational changes occur, leading to the release of mature mRNA (complex I). Final conformation changes then take place, resulting in the release of the lariat intron and dissembly of the spliceosome (or complex I).
Protein Factors Facilitate and Modulate Spliceosomal Conformational Changes
It has long been proposed that the rearrangements of RNA-RNA interactions and/or conformational changes are orchestrated by spliceosomal protein factors (7). Indeed, multiple such protein factors have been identified, and many of them are members of the family of DExH/D box ATPases, which can either directly promote the unwinding of RNA-RNA duplexes (also referred to as unwindases) or disrupt RNA-protein interactions (or even protein-protein interactions) (91). These spliceosomal ATPases function in a substrate-specific way, such that they each promote a specific conformational change (or RNA-RNA interaction) at various stages along the pathway of spliceosome assembly and catalysis. For instance, Prp5 is an RNA-dependent ATPase that bridges U1 and U2 snRNPs at early times during spliceosome assembly, promoting stable interaction of U2 with the intron branch site (40, 92) and thus promoting complex A formation. Also, there is evidence that the incorporation of U2 snRNP into the spliceosome is facilitated by Sub2/UAP56 (U2AF-associated protein), another RNA-dependent spliceosomal ATPase that acts through disruption of an interaction between U2AF and the branch site (93). Brr2, a U5 snRNP-associated protein and also an RNA-dependent ATPase, promotes the dissociation of U6 from U4 (94) in the spliceosome (presumably soon after complex B1 formation). In vivo analysis in S. cerevisiae suggests that the spliceosomal ATPase Prp28 may be actively disrupting the interaction between U1-C and the 5' splice site so as to allow for the exchange of U1 for U6 (95, 96), which is part of the rearrangement occurring during the transition from complex B1 to complex B2. The spliceosomal ATPase Prp2, on the other hand, promotes the first step of splicing (97-101), presumably also through rearranging the RNA-RNA interactions required for catalysis. The spliceosomal ATPase Prp16 promotes the conformational changes required for the second step of splicing (transition from complex B2 to complex C1) (90, 102-105). Prp22, a splicesomal ATPase as well, facilitates the conformational changes required for the release of mRNA (106, 107), promoting the transition from complex C1 to complex I. Finally, another spliceosomal ATPase Prp43 orchestrates the conformational changes required for the release of the lariat intron (108).
Besides DExH/D box ATPases, Snu114, a component of U5 snRNP and a GTPase, is also required for splicing, presumably through promoting the rearrangement between Prp8 and Snu114 itself, thus leading to the release of U1 and U4 before the first step of splicing (109). In addition to the ATPases/GTPase, several other spliceosomal proteins facilitate the rearrangements as well. Included in these proteins are the U5 snRNP-associated proteins p116 and p220 (Prp8p in yeast), which are thought to play a regulatory role in the unwinding of U4/U6 (110). Yeast genetic analyses indicate that Prp8 also plays a role in modulating the function of the splicesomal ATPases, Prp16 (90, 105) and Prp22 (111). Likewise, Isy1, a component of Prp NineTeen Complex (NTC), plays a role in facilitating the function of Prp16 (112). Similar roles can be assigned to U1 C and Cus2, which are implicated in modulating the functions of Prp28 and Prp5, respectively (96, 113). Importantly, the spliceosomal ATPases and their modulators are not only the key factors in orchestrating the conformational changes during spliceosome assembly and catalysis, but also play important roles in maintaining the fidelity of splicing. In this regard, it has been proposed that incorrect conformations (e.g., those caused by mutations in pre-mRNA and/or other spliceosomal factors) are discarded through these conformational changes catalyzed by the splicesomal ATPases and their modulators (90, 105, 114-116).
Prefabricated Spliceosome?
In contrast to the stepwise spliceosome assembly model, recent observations have indicated that the spliceosome exists as a preformed complex that engages the pre-mRNA as such 117-121). The strongest evidence for this model comes from the S. cerevisiae system from which a 45S snRNP complex was isolated that contained all five snRNPs (121). This penta-snRNP complex contains nearly all known U1, U2, and U4/U6-U5 snRNP proteins as well as non-snRNP splicing factors. The complex has also been shown to function in yeast cell extracts depleted of endogenous RNAs by nuclease digestion. Importantly, while assembling onto the pre-mRNA, the snRNA constituents of the penta-snRNP do not exchange with endogenous snRNPs. Consistent with the idea of a preassembled spliceosome, it has also been reported that both U2 and the U4/U6-U5 tri-snRNP function before the formation of complex A (122). Furthermore, recent analysis of splicing complex formation in HeLa nuclear extracts argues that a preassembled 200S RNP complex containing all snRNP components can assemble onto a short RNA containing a 5' splice site (123). However, using chromatin im- munoprecipitationmore (ChIP) analysis, the Neugebauer group (124, 125) and the Rosbash laboratory (126, 127) have more recently suggested that the recruitment of spliceosomal snRNPs to nascent pre-mRNA (in vivo spliceosome assembly) in yeast occurs via a stepwise assembly pathway, which is similar to that observed in vitro (see above, Spliceosome Assembly Pathway). Thus, it remains controversial as to whether spliceosome assembly is a one-step process (the prefabricated penta-snRNP complex assembles onto pre-mRNA) or it proceeds in a stepwise fashion. Further study is necessary to clarify this important issue.
Is Splicing an RNA-Catalyzed Reaction?
It has long been suspected that the two trans-esterification reactions to remove the introns from a pre-mRNA are catalyzed by the RNA constituents in the spliceosome (128). This idea is bolstered by the fact that U snRNAs in the spliceosome do form functional structures (the extended U6 intramolecular stem in complex B2 and the U6 intramolecular stem plus U2-U6 Helix Ib in complex C1; see Fig. 1) resembling domain V of group II introns, some of which are self-spliced via the two-step trans-esterification pathway identical to that of spliceosome-catalyzed pre-mRNA splicing (see above). Domain V of group II introns constitutes the catalytic center for catalysis during self-splicing (129-131).
Over the past decade, a growing body of evidence has suggested that pre-mRNA splicing in the spliceosome may indeed be catalyzed by its RNA constituents. For instance, Sontheimer et al. have shown that the first catalytic step of splicing occurs through a metal-ion-dependent pathway (132), an observation consistent with the two metal-ion model proposed for the spliceosome active site(s) (133). Later, using sulphur substitution followed by manganese suppression, Lin and colleagues have demonstrated that, through metal ion coordination, U6 plays a critical role in the catalysis reaction (134). Most recently, Valadkhan and Manley carried out an even more direct experiment to address this issue (135). In that experiment, they used only three short RNA oligonucleotides, corresponding to U2, U6 (designed according to U2-U6 Helices I, II, III, and the U6 intramolecular stem, see Figs. 1 and 3) and the branch site, respectively, and no proteins were included. Remarkably, the U2 and U6 oligonucleotides do form the functional structure for the first step of splicing (U2-U6 Helix I plus the extended intramolecular stem of U6), leading to the production of X-RNA, a product generated by a splicing-like reaction by which the branch point adenosine in the branch site oligonucleotide attacks a phosphate in the U6 oligonucleotide (Fig. 3). Taken together, the evidence accumulated thus far strongly supports the RNA-catalysis model for the spliceosome.
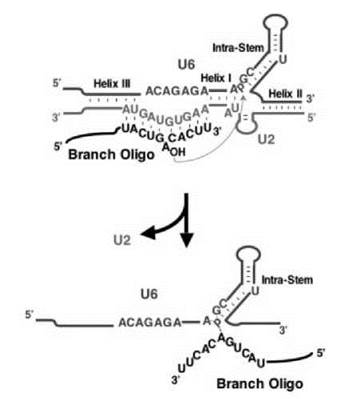
Figure 3. Protein-independent catalysis. According to the catalytic center of the spliceosome (see Fig. 1), three short RNA oligonucleotides, corresponding to an important fragment of U2 (green), an important fragment of U6 (brown), and the intron branch site (black), are designed. Indeed, they fold, in the absence of protein, into a structure that resembles the catalytic center of the spliceosome (see Fig. 1, the RNA structure in complex B2) or domain V of the self-splicing Group II intron. A splicing-like reaction occurs by which the branch point adenosine nucleophilically attacks the phosphate located between A and G at the base of the extended U6 intramolecular stem, generating a splicing-related product as indicated. U2-U6 Helices I, II, and III, the extended U6 intramolecular stem, and the base-pairing between the U2 branch recognition sequence and the branch site are shown. (Modified from Ref. 135.)
A Parallel Spliceosome
In high eukaryotes, there also exists a minor population of introns containing distinct consensus sequences at their 5' and 3’ ends (136). The removal of these introns takes place in an analogous spliceosome containing a different set of U snRNPs (U11, U12, U5, U4atac, and U6atac) (137). Although initially dubbed AT-AC introns based on their termini, extensive genomic database surveys have revealed that the standard GT-AG terminal sequences are more prevalent (138, 139). Instead, what appears to distinguish this class of introns are longer and more constrained consensus sequences at the 5' end of the intron and the branch site, as well as the absence of a polypyrimidine tract upstream of the 3' splice site (138, 139). The splicing machinery responsible for the removal of these introns is of much lower abundance (~104 copies per cell) relative to components of the major spliceosome, which is compatible with the low frequency in which these introns appear in the genome (~1/300 human introns) (140).
Remarkably, U11, U12, U4atac, and U6atac form structures that are almost identical to their counterparts in the major spliceosome, namely U1, U2, U4, and U6, respectively, despite the fact that the two sets of snRNAs are quite different in primary sequences (136, 137). Equally strikingly, the network of RNA-RNA interactions detected in the major spliceosome also exists in the U12-dependent spliceosome, further validating the importance of these dynamic interactions during spliceosome assembly and splicing (136, 137).
The main mechanistic differences between the two spliceosomes occur at the stage of intron recognition, rather than catalysis. Indeed, recognition of the 5' splice site and the branch site occurs simultaneously by the U11-U12 di-snRNP (141). Furthermore, there is a requirement among U12-dependent introns for 5' exon sequences to form U6atac-5' splice site interactions (142). Lastly, more constrained consensus sequences, as well as the lack of a polypyrimidine tract, suggest that the assembly of this spliceosome is more dependent on snRNA-based interactions than assembly of the major spliceosome (140, 143).
Acknowledgments
We thank our colleagues in the Yu lab for inspiration. Our work was supported by grant GM62937 (to Y.-T. Yu) from the National Institute of Health.
References
1. Chow LT, Gelinas RE, Broker TR, Roberts RJ. An amazing sequence arrangement at the 5' ends of adenovirus 2 messenger RNA. Cell 1977; 12:1-8.
2. Berget SM, Moore C, Sharp PA. Spliced segments at the 5' terminus of adenovirus 2 late mRNA. Proc. Natl. Acad. Sci. U.S.A. 1977; 74:3171-3175.
3. Gilbert W. Why genes in pieces? Nature 1978; 271:501.
4. Tilghman SM, Curtis PJ, Tiemeier DC, Leder P, Weissmann C. The intervening sequence of a mouse beta-globin gene is transcribed within the 15S beta-globin mRNA precursor. Proc. Natl. Acad. Sci. U.S.A. 1978; 75:1309-1313.
5. Burge CB, Tuschl T, Sharp PA. Splicing of precursors to mRNAs by the spliceosome. In: The RNA World. Gesteland RF, Cech TR, Atkins JF, eds. 1999. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
6. Yu YT, Scharl EC, Smith CM, Steitz JA. The growing world of small nuclear ribonucleoproteins. In: The RNA World. Gesteland RF, Cech TR, Atkins JF, eds. 1999. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
7. Staley JP, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 1998; 92:315-326.
8. Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell. 2003; 12:5-14.
9. Nilsen TW. RNA-RNA interactions in the spliceosome: unraveling the ties that bind. Cell 1994; 78:1-4.
10. Nilsen TW. RNA-RNA interactions in nuclear pre-mRNA splicing. In: RNA Structure and Function. Simons RW, Grunberg- Manago M, eds. 1998. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
11. Madhani HD, Guthrie C. Dynamic RNA-RNA interactions in the spliceosome. Annu. Rev. Genet. 1994; 28:1-26.
12. Mount SM. A catalogue of splice junction sequences. Nucleic Acids Res. 1982; 10:459-472.
13. Green MR. Pre-mRNA splicing. Annu. Rev. Genet. 1986; 20:671-708.
14. Hodnett JL, Busch H. Isolation and characterization of uridylic acid-rich 7S ribonucleic acid of rat liver nuclei. J. Biol. Chem. 1968; 243:6334-6342.
15. Weinberg RA, Penman S. Small molecular weight monodisperse nuclear RNA. J. Mol. Biol. 1968; 38:289-304.
16. Lerner MR, Boyle JA, Mount SM, Wolin SL, Steitz JA. Are snRNPs involved in splicing? Nature 1980; 283:220-224.
17. Rogers J, Wall R. A mechanism for RNA splicing. Proc. Natl. Acad. Sci. U.S.A. 1980; 77:1877-1879.
18. Mount SM, Steitz JA. Sequence of U1 RNA from Drosophila melanogaster: implications for U1 secondary structure and possible involvement in splicing. Nucleic Acids Res. 1981; 9:6351-6368.
19. Mount SM, Pettersson I, Hinterberger M, Karmas A, Steitz JA. The U1 small nuclear RNA-protein complex selectively binds a 5' splice site in vitro. Cell 1983; 33:509-518.
20. Kramer A, Keller W, Appel B, Luhrmann R. The 5' terminus of the RNA moiety of U1 small nuclear ribonucleoprotein particles is required for the splicing of messenger RNA precursors. Cell 1984; 38:299-307.
21. Zhuang Y, Weiner AM. A compensatory base change in U1 snRNA suppresses a 5' splice site mutation. Cell 1986; 46:827-835.
22. Wassarman DA, Steitz JA. Interactions of small nuclear RNA’s with precursor messenger RNA during in vitro splicing. Science 1992; 257:1918-1925.
23. Legrain P, Seraphin B, Rosbash M. Early commitment of yeast pre-mRNA to the spliceosome pathway. Mol. Cell Biol. 1988; 8:3755-3760.
24. Seraphin B, Rosbash M. Identification of functional U1 snRNA-pre-mRNA complexes committed to spliceosome assembly and splicing. Cell 1989; 59:349-358.
25. Ruby SW, Abelson J. An early hierarchic role of U1 small nuclear ribonucleoprotein in spliceosome assembly. Science 1988; 242:1028-1035.
26. Seraphin B, Rosbash M. The yeast branchpoint sequence is not required for the formation of a stable U1 snRNA-pre-mRNA complex and is recognized in the absence of U2 snRNA. EMBO J. 1991; 10:1209-1216.
27. Michaud S, Reed R. An ATP-independent complex commits pre-mRNA to the mammalian spliceosome assembly pathway. Genes Dev. 1991; 5:2534-2546.
28. Bindereif A, Green MR. An ordered pathway of snRNP binding during mammalian pre-mRNA splicing complex assembly. EMBO J. 1987; 6:2415-2424.
29. Steitz JA, Black DL, Gerke V, Parker KA, Kramer A, Frendewey D, Keller W. Functions of the abundant U-snRNPs. In: Small Nuclear Ribonucleoprotein Particles. Birnstiel ML, ed. 1988. Springer-Verlag, Berlin.
30. Zhuang Y, Weiner AM. A compensatory base change in human U2 snRNA can suppress a branch site mutation. Genes Dev. 1989; 3:1545-1552.
31. Zhuang YA, Goldstein AM, Weiner AM. UACUAAC is the preferred branch site for mammalian mRNA splicing. Proc. Natl. Acad. Sci. U.S.A. 1989; 86:2752-2756.
32. Parker R, Siliciano PG, Guthrie C. Recognition of the TACTAAC box during mRNA splicing in yeast involves base pairing to the U2-like snRNA. Cell 1987; 49:229-239.
33. Black DL, Chabot B, Steitz JA. U2 as well as U1 small nuclear ribonucleoproteins are involved in premessenger RNA splicing. Cell 1985; 42:737-750.
34. Krainer AR, Maniatis T. Multiple factors including the small nuclear ribonucleoproteins U1 and U2 are necessary for pre-mRNA splicing in vitro. Cell 1985; 42:725-736.
35. Konarska MM, Sharp PA. Electrophoretic separation of complexes involved in the splicing of precursors to mRNAs. Cell 1986; 46:845-855.
36. Will CL, Luhrmann R. Spliceosome structure and function. In: The RNA World. Gesteland RF, Cech TR, Atkins JF, eds. 2006. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
37. Will CL, Luhrmann R. Protein functions in pre-mRNA splicing. Curr. Opin. Cell Biol. 1997; 9:320-328.
38. Fu XD. The superfamily of arginine/serine-rich splicing factors. RNA 1995; 1:663-680.
39. Reed R. Initial splice-site recognition and pairing during pre-mRNA splicing. Curr. Opin. Genet. Dev. 1996; 6:215-220.
40. Xu YZ, Newnham CM, Kameoka S, Huang T, Konarska MM, Query CC. Prp5 bridges U1 and U2 snRNPs and enables stable U2 snRNP association with intron RNA. EMBO J. 2004; 23:376-385.
41. Singh R, Valcarcel J, Green MR. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science 1995; 268:1173-1176.
42. Wu S, Romfo CM, Nilsen TW, Green MR. Functional recognition of the 3' splice site AG by the splicing factor U2AF35. Nature 1999; 402:832-835.
43. Zorio DA, Blumenthal T. Both subunits of U2AF recognize the 3' splice site in Caenorhabditis elegans. Nature 1999; 402:835-838.
44. Merendino L, Guth S, Bilbao D, Martinez C, Valcarcel J. Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3' splice site AG. Nature 1999; 402:838-841.
45. Chua K, Reed R. The RNA splicing factor hSlu7 is required for correct 3' splice-site choice. Nature 1999; 402:207-210.
46. Frank D, Guthrie C. An essential splicing factor, SLU7, mediates 3' splice site choice in yeast. Genes Dev. 1992; 6:2112-2124.
47. Padgett RA, Konarska MM, Grabowski PJ, Hardy SF, Sharp PA. Lariat RNA’s as intermediates and products in the splicing of messenger RNA precursors. Science 1984; 225:898-903.
48. Ruskin B, Krainer AR, Maniatis T, Green MR. Excision of an intact intron as a novel lariat structure during pre-mRNA splicing in vitro. Cell 1984; 38:317-331.
49. Domdey H, Apostol B, Lin RJ, Newman A, Brody E, Abelson J. Lariat structures are in vivo intermediates in yeast pre-mRNA splicing. Cell 1984; 39:611-621.
50. Wallace JC, Edmonds M. Polyadenylylated nuclear RNA contains branches. Proc. Natl. Acad. Sci. U.S.A. 1983; 80:950-954.
51. Lin RJ, Newman AJ, Cheng SC, Abelson J. Yeast mRNA splicing in vitro. J. Biol. Chem. 1985; 260:14780-14792.
52. Brody E, Abelson J. The “spliceosome”: yeast pre-messenger RNA associates with a 40S complex in a splicing-dependent reaction. Science 1985; 228:963-967.
53. Grabowski PJ, Seiler SR, Sharp PA. A multicomponent complex is involved in the splicing of messenger RNA precursors. Cell 1985; 42:345-353.
54. Frendewey D, Keller W. Stepwise assembly of a pre-mRNA splicing complex requires U-snRNPs and specific intron sequences. Cell 1985; 42:355-367.
55. Bindereif A, Green MR. Ribonucleoprotein complex formation during pre-mRNA splicing in vitro. Mol. Cell Biol. 1986; 6:2582-2592.
56. Perkins KK, Furneaux HM, Hurwitz J. RNA splicing products formed with isolated fractions from HeLa cells are associated with fast-sedimenting complexes. Proc. Natl. Acad. Sci. U.S.A. 1986; 83:887-891.
57. Kaltwasser G, Spitzer SG, Goldenberg CJ. Assembly in an in vitro splicing reaction of a mouse insulin messenger RNA precursor into a 60-40S ribonucleoprotein complex. Nucleic Acids Res. 1986; 14:3687-3701.
58. Pikielny CW, Rymond BC, Rosbash M. Electrophoresis of ribonucleoproteins reveals an ordered assembly pathway of yeast splicing complexes. Nature 1986; 324:341-345.
59. Pikielny CW, Rosbash M. Specific small nuclear RNAs are associated with yeast spliceosomes. Cell 1986; 45:869-877.
60. Frendewey D, Kramer A, Keller W. Different small nuclear ribonucleoprotein particles are involved in different steps of splicing complex formation. Cold Spring Harb. Symp. Quant. Biol. 1987; 52:287-298.
61. Cheng SC, Abelson J. Spliceosome assembly in yeast. Genes Dev. 1987; 1:1014-1027.
62. Konarska, M, Sharp PA. Interactions between small nuclear ribonucleoprotein particles in formation of spliceosomes. Cell 1987; 49:763-774.
63. Grabowski PJ, Sharp PA. Affinity chromatography of splicing complexes: U2, U5, and U4 + U6 small nuclear ribonucleoprotein particles in the spliceosome. Science 1986; 233:1294-1299.
64. Reed R. Protein composition of mammalian spliceosomes assembled in vitro. Proc. Natl. Acad. Sci. U.S.A. 1990; 87:8031-8035.
65. Bennett M, Michaud S, Kingston J, Reed R. Protein components specifically associated with prespliceosome and spliceosome complexes. Genes Dev. 1992; 6:1986-2000.
66. Moore MJ, Query CC, Sharp PA. Splicing of precursors to mRNAs by the spliceosome. In: The RNA World. Gesteland RF, Atkins JF, eds. 1993. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
67. Ruby SW, Abelson J. Pre-mRNA splicing in yeast. Trends Genet. 1991; 7:79-85.
68. Dreyfuss G, Swanson MS, Pinol-Roma S. The composition, structure, and organization of proteins in heterogeneous nuclear ribonucleoprotein complexes. In: The Eukaryotic Nucleus: Structure and Function. Strauss P, Wilson S, eds. 1990. Telford Press, Caldwell, NJ.
69. Lamond AI, Konarska MM, Grabowski PJ, Sharp PA. Spliceosome assembly involves the binding and release of U4 small nuclear ribonucleoprotein. Proc. Natl. Acad. Sci. U.S.A. 1988; 85:411-415.
70. Seraphin B, Abovich N, Rosbash M. Genetic depletion indicates a late role for U5 snRNP during in vitro spliceosome assembly. Nucleic Acids Res. 1991; 19:3857-3860.
71. Lamm GM, Blencowe BJ, Sproat BS, Iribarren AM, Ryder U, Lamond AI. Antisense probes containing 2-aminoadenosine allow efficient depletion of U5 snRNP from HeLa splicing extracts. Nucleic Acids Res. 1991; 19:3193-3198.
72. Utans U, Behrens SE, Luhrmann R, Kole R, Kramer A. A splicing factor that is inactivated during in vivo heat shock is functionally equivalent to the U4/U6.U5 triple snRNP-specific proteins. Genes Dev. 1992; 6:631-641.
73. Banroques J, Abelson JN. PRP4: a protein of the yeast U4/U6 small nuclear ribonucleoprotein particle. Mol. Cell Biol. 1989; 9:3710-3719.
74. Behrens SE, Luhrmann R. Immunoaffinity purification of a U4/U6.U5 tri-snRNP from human cells. Genes Dev. 1991; 5:1439-1452.
75. Wyatt JR, Sontheimer EJ, Steitz JA. Site-specific cross-linking of mammalian U5 snRNP to the 5’ splice site before the first step of pre-mRNA splicing. Genes Dev. 1992; 6:2542-2553.
76. Sontheimer EJ, Steitz JA. The U5 and U6 small nuclear RNAs as active site components of the spliceosome. Science 1993; 262:1989-1996.
77. Newman AJ, Teigelkamp S, Beggs JD. snRNA interactions at 5’ and 3’ splice sites monitored by photoactivated crosslinking in yeast spliceosomes. RNA 1995; 1:968-980.
78. Hausner TP, Giglio LM, Weiner AM. Evidence for base-pairing between mammalian U2 and U6 small nuclear ribonucleoprotein particles. Genes Dev. 1990; 4:2146-2156.
79. Sawa H, Abelson J. Evidence for a base-pairing interaction between U6 small nuclear RNA and 5’ splice site during the splicing reaction in yeast. Proc. Natl. Acad. Sci. U.S.A. 1992; 89:11269-11273.
80. Madhani HD, Guthrie C. Randomization-selection analysis of snRNAs in vivo: evidence for a tertiary interaction in the spliceosome. Genes Dev. 1994; 8:1071-1086.
81. Madhani HD, Guthrie C. A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell 1992; 71:803-817.
82. Datta B, Weiner AM. Genetic evidence for base pairing between U2 and U6 snRNA in mammalian mRNA splicing. Nature 1991; 352:821-824.
83. Newman AJ, Norman C. U5 snRNA interacts with exon sequences at 5’ and 3’ splice sites. Cell 1992; 68:743-754.
84. Newman A, Norman C. Mutations in yeast U5 snRNA alter the specificity of 5’ splice-site cleavage. Cell 1991; 65:115-123.
85. Sun JS, Manley JL. A novel U2-U6 snRNA structure is necessary for mammalian mRNA splicing. Genes Dev. 1995; 9:843-854.
86. Wu JA, Manley JL. Base pairing between U2 and U6 snRNAs is necessary for splicing of a mammalian pre-mRNA. Nature 1991; 352:818-821.
87. Cortes JJ, Sontheimer EJ, Seiwert SD, Steitz JA. Mutations in the conserved loop of human U5 snRNA generate use of novel cryptic 5’ splice sites in vivo. EMBO J. 1993; 12:5181-5189.
88. Lesser CF, Guthrie C. Mutations in U6 snRNA that alter splice site specificity: implications for the active site. Science 1993; 262:1982-1988.
89. Sashital DG, Cornilescu G, McManus CJ, Brow DA, Butcher SE. U2-U6 RNA folding reveals a group II intron-like domain and a four-helix junction. Nat. Struct. Mol. Biol. 2004; 11:1237-1242.
90. Konarska MM, Vilardell J, Query CC. Repositioning of the reaction intermediate within the catalytic center of the spliceosome. Mol. Cell 2006; 21:543-553.
91. Schwer B. A new twist on RNA helicases: DExH/D box proteins as RNPases. Nat. Struct. Biol. 2001; 8:113-116.
92. O’Day CL, Dalbadie-McFarland G, Abelson J. The Saccharomyces cerevisiae Prp5 protein has RNA-dependent ATPase activity with specificity for U2 small nuclear RNA. J. Biol. Chem. 1996; 271:33261-33267.
93. Kistler AL, Guthrie C. Deletion of MUD2, the yeast homolog of U2AF65, can bypass the requirement for sub2, an essential spliceosomal ATPase. Genes Dev. 2001; 15:42-49.
94. Raghunathan PL, Guthrie C. RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Curr. Biol. 1998; 8:847-855.
95. Staley JP, Guthrie C. An RNA switch at the 5’ splice site requires ATP and the DEAD box protein Prp28p. Mol. Cell 1999; 3:55-64.
96. Chen JY, Stands L, Staley JP, Jackups RR Jr, Latus LJ, Chang TH. Specific alterations of U1-C protein or U1 small nuclear RNA can eliminate the requirement of Prp28p, an essential DEAD box splicing factor. Mol. Cell 2001; 7:227-232.
97. King DS, Beggs JD. Interactions of PRP2 protein with pre-mRNA splicing complexes in Saccharomyces cerevisiae. Nucleic Acids Res. 1990; 18:6559-6564.
98. Kim SH, Lin RJ. Pre-mRNA splicing within an assembled yeast spliceosome requires an RNA-dependent ATPase and ATP hydrolysis. Proc. Natl. Acad. Sci. U.S.A. 1993; 90:888-892.
99. Edwalds-Gilbert G, Kim DH, Silverman E, Lin RJ. Definition of a spliceosome interaction domain in yeast Prp2 ATPase. RNA 2004; 10:210-220.
100. Plumpton M, McGarvey M, Beggs JD. A dominant negative mutation in the conserved RNA helicase motif ‘SAT’ causes splicing factor PRP2 to stall in spliceosomes. EMBO J. 1994; 13:879-887.
101. Kim SH, Smith J, Claude A, Lin RJ. The purified yeast pre-mRNA splicing factor PRP2 is an RNA-dependent NTPase. EMBO J. 1992; 11:2319-2326.
102. Schwer B, Guthrie C. A conformational rearrangement in the spliceosome is dependent on PRP16 and ATP hydrolysis. EMBO J. 1992; 11:5033-5039.
103. Schwer B, Guthrie C. A dominant negative mutation in a spliceosomal ATPase affects ATP hydrolysis but not binding to the spliceosome. Mol. Cell Biol. 1992; 12:3540-3547.
104. Schwer B, Guthrie C. PRP16 is an RNA-dependent ATPase that interacts transiently with the spliceosome. Nature 1991; 349:494-499.
105. Query CC, Konarska MM. Suppression of multiple substrate mutations by spliceosomal prp8 alleles suggests functional correlations with ribosomal ambiguity mutants. Mol. Cell 2004; 14: 343-354.
106. Schwer B, Gross CH. Prp22, a DExH-box RNA helicase, plays two distinct roles in yeast pre-mRNA splicing. EMBO J. 1998; 17:2086-2094.
107. Honig A, Auboeuf D, Parker MM, O’Malley BW, Berget SM. Regulation of alternative splicing by the ATP-dependent DEAD-box RNA helicase p72. Mol. Cell Biol. 2002; 22:5698-5707.
108. Martin A, Schneider S, Schwer B. Prp43 is an essential RNA-dependent ATPase required for release of lariat-intron from the spliceosome. J. Biol. Chem. 2002; 277:17743-17750.
109. Brenner TJ, Guthrie C. Assembly of Snu114 into U5 snRNP requires Prp8 and a functional GTPase domain. RNA 2006; 12:862-871.
110. Kuhn AN, Reichl EM, Brow DA. Distinct domains of splicing factor Prp8 mediate different aspects of spliceosome activation. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:9145-9149.
111. Schneider S, Campodonico E, Schwer B. Motifs IV and V in the DEAH box splicing factor Prp22 are important for RNA unwinding, and helicase-defective Prp22 mutants are suppressed by Prp8. J. Biol. Chem. 2004; 279:8617-8626.
112. Villa T, Guthrie C. The Isy1p component of the NineTeen complex interacts with the ATPase Prp16p to regulate the fidelity of pre-mRNA splicing. Genes Dev. 2005; 19:1894-1904.
113. Perriman R, Barta I, Voeltz GK, Abelson J, Ares M Jr. ATP requirement for Prp5p function is determined by Cus2p and the structure of U2 small nuclear RNA. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:13857-13862.
114. Konarska MM, Query CC. Insights into the mechanisms of splicing: more lessons from the ribosome. Genes Dev. 2005; 19:2255-2260.
115. Query CC, Konarska MM. Splicing fidelity revisited. Nat. Struct. Mol. Biol. 2006; 13:472-474.
116. Mayas RM, Maita H, Staley JP. Exon ligation is proofread by the DExD/H-box ATPase Prp22p. Nat. Struct. Mol. Biol. 2006; 13:482-490.
117. Gottschalk A, Neubauer G, Banroques J, Mann M, Luhrmann R, Fabrizio P. Identification by mass spectrometry and functional analysis of novel proteins of the yeast U4/U6.U5 tri-snRNP. EMBO J. 1999; 18:4535-4548.
118. Ohi MD, Link AJ, Ren L, Jennings JL, McDonald WH, Gould KL. Proteomics analysis reveals stable multiprotein complexes in both fission and budding yeasts containing Myb-related Cdc5p/Cef1p, novel pre-mRNA splicing factors, and snRNAs. Mol. Cell Biol. 2002; 22:2011-2024.
119. Wang Q, Hobbs K, Lynn B, Rymond BC. The Clf1p splicing factor promotes spliceosome assembly through N-terminal tetratricopeptide repeat contacts. J. Biol. Chem. 2003; 278:7875-7883.
120. Huang T, Vilardell J, Query CC. Pre-spliceosome formation in S.pombe requires a stable complex of SF1-U2AF59-U2AF23. EMBO J. 2002; 21:5516-5526.
121. Stevens SW, Ryan DE, Ge HY, Moore RE, Young MK, Lee TD, Abelson J. Composition and functional characterization of the yeast spliceosomal penta-snRNP. Mol. Cell 2002; 9:31-44.
122. Maroney PA, Romfo CM, Nilsen TW. Functional recognition of 5’ splice site by U4/U6.U5 tri-snRNP defines a novel ATP-dependent step in early spliceosome assembly. Mol. Cell 2000; 6:317-328.
123. Malca H, Shomron N, Ast G. The U1 snRNP base pairs with the 5’ splice site within a penta-snRNP complex. Mol. Cell Biol. 2003; 23:3442-3455.
124. Kotovic KM, Lockshon D, Boric L, Neugebauer KM. Cotranscriptional recruitment of the U1 snRNP to intron-containing genes in yeast. Mol. Cell Biol. 2003; 23:5768-5779.
125. Gornemann J, Kotovic KM, Hujer K, Neugebauer KM. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol. Cell 2005; 19:53-63.
126. Lacadie SA, Rosbash M. Cotranscriptional spliceosome assembly dynamics and the role of U1 snRNA:5’ss base pairing in yeast. Mol. Cell 2005; 19:65-75.
127. Tardiff DF, Rosbash M. Arrested yeast splicing complexes indicate stepwise snRNP recruitment during in vivo spliceosome assembly. RNA 2006; 12:968-979.
128. Sharp PA. Five easy pieces. Science 1991; 254:663.
129. Jarrell KA, Dietrich RC, Perlman PS. Group II intron domain 5 facilitates a trans-splicing reaction. Mol. Cell Biol. 1988; 8:2361-2366.
130. Koch JL, Boulanger SC, Dib-Hajj SD, Hebbar SK, Perlman PS. Group II introns deleted for multiple substructures retain self-splicing activity. Mol. Cell Biol. 1992; 12:1950-1958.
131. Michel F, Umesono K, Ozeki H. Comparative and functional anatomy of group II catalytic introns—a review. Gene 1989; 82:5-30.
132. Sontheimer EJ, Sun S, Piccirilli JA. Metal ion catalysis during splicing of premessenger RNA. Nature 1997; 388:801-805.
133. Steitz TA, Steitz JA. A general two-metal-ion mechanism for catalytic RNA. Proc. Natl. Acad. Sci. U.S.A. 1993; 90:6498-6502.
134. Yean SL, Wuenschell G, Termini J, Lin RJ. Metal-ion coordination by U6 small nuclear RNA contributes to catalysis in the spliceosome. Nature 2000; 408:881-884.
135. Valadkhan S, Manley JL. Splicing-related catalysis by protein-free snRNAs. Nature 2001; 413:701-707.
136. Tarn WY, Steitz JA. Pre-mRNA splicing: the discovery of a new spliceosome doubles the challenge. Trends Biochem. Sci. 1997; 22:132-137.
137. Tarn WY, Steitz JA. Highly diverged U4 and U6 small nuclear RNAs required for splicing rare AT-AC introns. Science 1996; 273:1824-1832.
138. Dietrich RC, Incorvaia R, Padgett RA. Terminal intron dinucleotide sequences do not distinguish between U2- and U12-dependent introns. Mol. Cell 1997; 1:151-160.
139. Sharp PA, Burge CB. Classification of introns: U2-type or U12-type. Cell 1997; 91:875-879.
140. Tycowski KT, Kolev NG, Conrad NK, Fok V, Steitz JA. The ever-growing world of small nuclear ribonucleoproteins. In: The RNA World. Gesteland RF, Cech TR, Atkins JF, eds. 2006. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
141. Frilander MJ, Steitz JA. Initial recognition of U12-dependent introns requires both U11/57 splice-site and U12/branchpoint interactions. Genes Dev. 1999; 13:851-863.
142. Frilander MJ, Steitz JA. Dynamic exchanges of RNA interactions leading to catalytic core formation in the U12-dependent spliceosome. Mol. Cell 2001; 7:217-226.
143. Patel AA, Steitz JA. Splicing double: insights from the second spliceosome. Nat. Rev. Mol. Cell Biol. 2003; 4:960-970.